Phylogenetic Tree Production
VerifiedAdded on 2022/08/25
|13
|1850
|38
AI Summary
Contribute Materials
Your contribution can guide someone’s learning journey. Share your
documents today.

Running head: BIOINFORMATICS
GLYCINE MAX
Name of the Student
Name of the University
Author Note
GLYCINE MAX
Name of the Student
Name of the University
Author Note
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.
1BIOINFORMATICS
Eleven steps will be followed to answer the questions as given below-
Answer 1
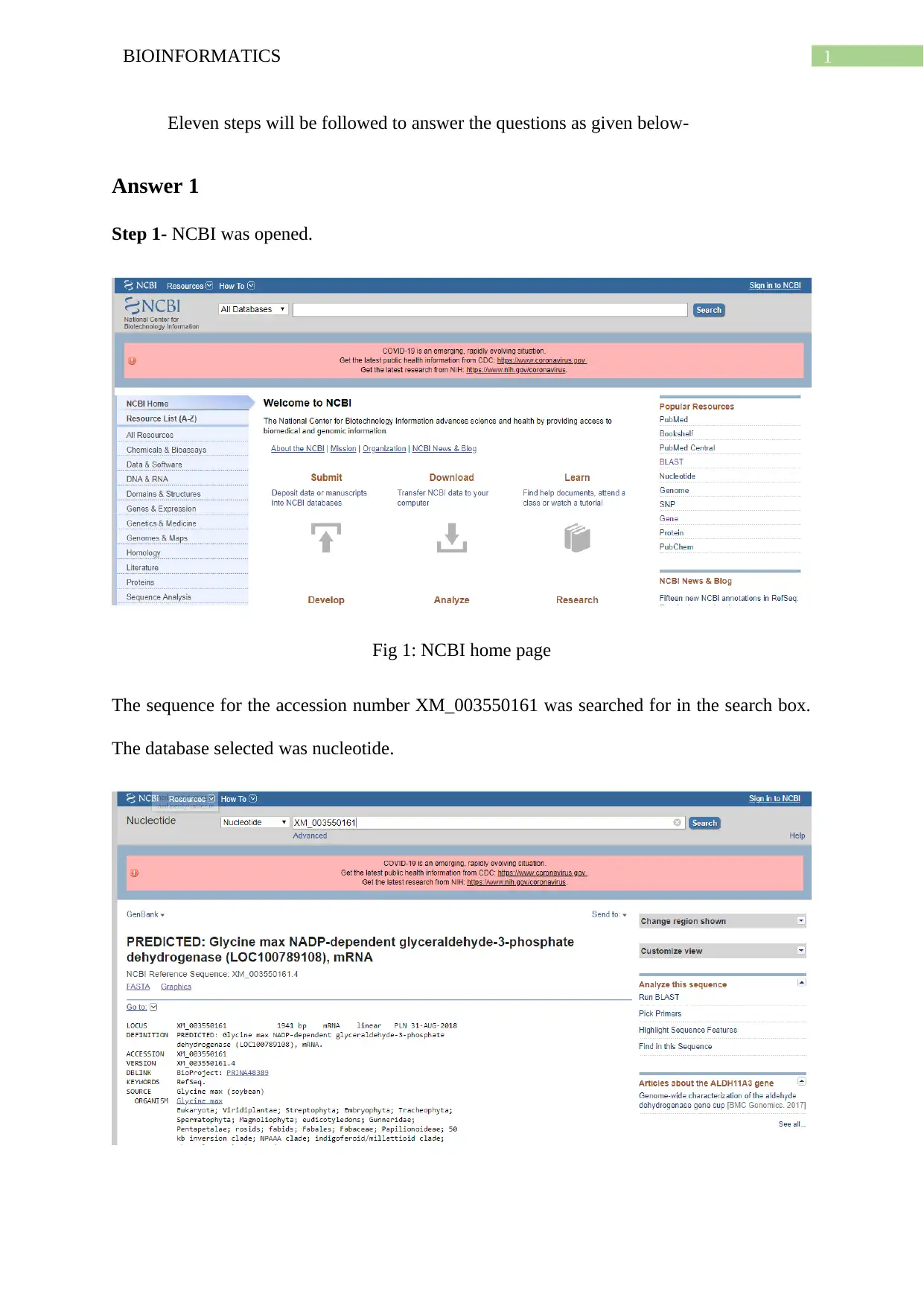
Step 1- NCBI was opened.
Fig 1: NCBI home page
The sequence for the accession number XM_003550161 was searched for in the search box.
The database selected was nucleotide.
Eleven steps will be followed to answer the questions as given below-
Answer 1
Step 1- NCBI was opened.
Fig 1: NCBI home page
The sequence for the accession number XM_003550161 was searched for in the search box.
The database selected was nucleotide.

2BIOINFORMATICS
Fig 2: NCBI results page
Thus, the name of the gene is “Glycine max NADP-dependent glyceraldehyde-3-
phosphate dehydrogenase (LOC100789108), mRNA”.
Step 2- Both the FASTA and genebank sequences were downloaded.
FASTA sequence:
>XM_003550161.4 PREDICTED: Glycine max NADP-dependent glyceraldehyde-3-
phosphate dehydrogenase (LOC100789108), mRNA
GACTAAGCACTAAGTATTAGCAACATTTATAGCTGTTCCTTGACCCTGTCTGAATCCACTTCAATAATCA
TGTTATACTAATAATAATCACCAACCACATAATCAACATCAATATATATATACTCCTAAGCCAAAGTACA
AACCTCAAACCAAATCGAATCACTTGTTAGTTCAACATGGCTGGCAGTGGCACCTTTGCAGAGATCATAG
ATGGTGATGTGTTCAAGTACTATGCACAAGGCCATTGGAACAAGTCCTCTTCTGGAAAATTCGTTCCCAT
CATCAATCCAACCACTAGGAAGACCCACTTCAAGGTCCAAGCTTGTACACAGAAAGAGGTTAATAGGGTC
ATGGAATCAGCAAAAACAGCTCAAAAATCATGGGCCAAGACTCCACTGTGGAAAAGAGCAGAGTTGCTTC
ACAAGGCTGCAGCAATACTGAAGGAGCACAAAGCACCTATTGCTGAGTGTCTTGTAAAGGAGATTGCAAA
GCCAGCTAAGGATGCTGTCACTGAGGTGATCAGATCAGGGGATTTGGTGTCATATTGTGCTGAGGAAGGG
GTGAGAATTCTTGGGGAAGGAAAATTTCTGGTCTCTGATAGTTTTCCGGGCAATGAAAGAACCAAATACT
GTCTCACTTCCAAGATTCCACTGGGAGTTGTTTTAGCCATTCCACCTTTCAACTATCCTGTTAATCTGGC
TGTTTCAAAGATTGCTCCAGCGCTTATTGCTGGAAACTCCATTGTTCTTAAACCCCCCACCCAGGGTGCT
GTTGCTGCACTACATATGGTGCATTGCTTTCATTTGGCTGGTTTCCCCGAAGGCCTTATTAGCTGTGTGA
CAGGAAAGGGTTCCGAGATTGGTGATTTTCTTACAATGCATCCGGGGGTGAACTGCATAAGTTTCACTGG
TGGGGACACTGGAATAGCAATATCAAAGAAGGCCGGCATGGTTCCTCTTCAAATGGAATTGGGTGGAAAA
GATGCCTGCATTGTTCTTGAAGATGCTGACTTGGATTTGGCAGCTGCTAACATTGTAAAAGGAGGCTTTT
CCTACAGTGGTCAAAGATGCACAGCTGTAAAGGTTGCTCTGGTCATGGAATCGGTTGCCAACACTCTTGT
AAAGAGAATCAATGATAAAATTGCAAAGTTAACTGTTGGACCTCCTGAGATTGACAGTGATGTCACACCA
GTTGTCACCGAATCCTCCGCTAACTTCATTGAAGGGTTGGTAATGGATGCTAAGGAGAAAGGAGCAACAT
TCTGCCAGGAATACGTAAGGGAGGGAAATCTCATTTGGCCATTGCTGTTGGATAATGTTCGTCCCGATAT
GAGGATAGCATGGGAGGAGCCATTTGGACCAGTTTTGCCAGTTATCAGGATAAATTCTGTTGAAGAAGGA
ATCCACCATTGTAATGCTAGCAATTTTGGCCTTCAGGGATGTGTCTTCACAAGAGACATTAACAAAGCAA
TGCTGATCAGTGATGCAATGGAGACAGGAACAGTTCAAATTAATTCAGCACCAGCTCGTGGACCTGATCA
CTTTCCTTTTCAGGGTCTGAAGGATAGTGGAATTGGTTCTCAAGGGATCACCAACAGTATTAACATGATG
ACAAAGGTTAAGACTACAATAATCAACTTGCCAGCACCTTCTTACACTATGGGATGAGAATAATGAGATG
TCCAAGAAACAGTTTAGTCACTCAAAGTAGTAGTAATCAGCATATAAAGTAGGCTTTTCCGATCTCCCAC
GTTACAGCTTAGCCCTATAGAGTTGAGTAAGGGACAGTCTTTGAGAGGGGGCTGCTTGCATACAAAAAAT
ATTTGTTGCAGTGCATGTGGATTAACTGTACTTTATGTTTCTATGGAGAAAAGGGTAATCACTATTTGTA
CAAGTAGGATATGAATCTTGAATAAATTATGGTTCTCCAACTTTACTTGAA
GeneBank sequence:
LOCUS XM_003550161 1941 bp mRNA linear PLN 31-AUG-
2018
DEFINITION PREDICTED: Glycine max NADP-dependent glyceraldehyde-3-
phosphate
dehydrogenase (LOC100789108), mRNA.
ACCESSION XM_003550161
VERSION XM_003550161.4
DBLINK BioProject: PRJNA48389
KEYWORDS RefSeq.
SOURCE Glycine max (soybean)
Fig 2: NCBI results page
Thus, the name of the gene is “Glycine max NADP-dependent glyceraldehyde-3-
phosphate dehydrogenase (LOC100789108), mRNA”.
Step 2- Both the FASTA and genebank sequences were downloaded.
FASTA sequence:
>XM_003550161.4 PREDICTED: Glycine max NADP-dependent glyceraldehyde-3-
phosphate dehydrogenase (LOC100789108), mRNA
GACTAAGCACTAAGTATTAGCAACATTTATAGCTGTTCCTTGACCCTGTCTGAATCCACTTCAATAATCA
TGTTATACTAATAATAATCACCAACCACATAATCAACATCAATATATATATACTCCTAAGCCAAAGTACA
AACCTCAAACCAAATCGAATCACTTGTTAGTTCAACATGGCTGGCAGTGGCACCTTTGCAGAGATCATAG
ATGGTGATGTGTTCAAGTACTATGCACAAGGCCATTGGAACAAGTCCTCTTCTGGAAAATTCGTTCCCAT
CATCAATCCAACCACTAGGAAGACCCACTTCAAGGTCCAAGCTTGTACACAGAAAGAGGTTAATAGGGTC
ATGGAATCAGCAAAAACAGCTCAAAAATCATGGGCCAAGACTCCACTGTGGAAAAGAGCAGAGTTGCTTC
ACAAGGCTGCAGCAATACTGAAGGAGCACAAAGCACCTATTGCTGAGTGTCTTGTAAAGGAGATTGCAAA
GCCAGCTAAGGATGCTGTCACTGAGGTGATCAGATCAGGGGATTTGGTGTCATATTGTGCTGAGGAAGGG
GTGAGAATTCTTGGGGAAGGAAAATTTCTGGTCTCTGATAGTTTTCCGGGCAATGAAAGAACCAAATACT
GTCTCACTTCCAAGATTCCACTGGGAGTTGTTTTAGCCATTCCACCTTTCAACTATCCTGTTAATCTGGC
TGTTTCAAAGATTGCTCCAGCGCTTATTGCTGGAAACTCCATTGTTCTTAAACCCCCCACCCAGGGTGCT
GTTGCTGCACTACATATGGTGCATTGCTTTCATTTGGCTGGTTTCCCCGAAGGCCTTATTAGCTGTGTGA
CAGGAAAGGGTTCCGAGATTGGTGATTTTCTTACAATGCATCCGGGGGTGAACTGCATAAGTTTCACTGG
TGGGGACACTGGAATAGCAATATCAAAGAAGGCCGGCATGGTTCCTCTTCAAATGGAATTGGGTGGAAAA
GATGCCTGCATTGTTCTTGAAGATGCTGACTTGGATTTGGCAGCTGCTAACATTGTAAAAGGAGGCTTTT
CCTACAGTGGTCAAAGATGCACAGCTGTAAAGGTTGCTCTGGTCATGGAATCGGTTGCCAACACTCTTGT
AAAGAGAATCAATGATAAAATTGCAAAGTTAACTGTTGGACCTCCTGAGATTGACAGTGATGTCACACCA
GTTGTCACCGAATCCTCCGCTAACTTCATTGAAGGGTTGGTAATGGATGCTAAGGAGAAAGGAGCAACAT
TCTGCCAGGAATACGTAAGGGAGGGAAATCTCATTTGGCCATTGCTGTTGGATAATGTTCGTCCCGATAT
GAGGATAGCATGGGAGGAGCCATTTGGACCAGTTTTGCCAGTTATCAGGATAAATTCTGTTGAAGAAGGA
ATCCACCATTGTAATGCTAGCAATTTTGGCCTTCAGGGATGTGTCTTCACAAGAGACATTAACAAAGCAA
TGCTGATCAGTGATGCAATGGAGACAGGAACAGTTCAAATTAATTCAGCACCAGCTCGTGGACCTGATCA
CTTTCCTTTTCAGGGTCTGAAGGATAGTGGAATTGGTTCTCAAGGGATCACCAACAGTATTAACATGATG
ACAAAGGTTAAGACTACAATAATCAACTTGCCAGCACCTTCTTACACTATGGGATGAGAATAATGAGATG
TCCAAGAAACAGTTTAGTCACTCAAAGTAGTAGTAATCAGCATATAAAGTAGGCTTTTCCGATCTCCCAC
GTTACAGCTTAGCCCTATAGAGTTGAGTAAGGGACAGTCTTTGAGAGGGGGCTGCTTGCATACAAAAAAT
ATTTGTTGCAGTGCATGTGGATTAACTGTACTTTATGTTTCTATGGAGAAAAGGGTAATCACTATTTGTA
CAAGTAGGATATGAATCTTGAATAAATTATGGTTCTCCAACTTTACTTGAA
GeneBank sequence:
LOCUS XM_003550161 1941 bp mRNA linear PLN 31-AUG-
2018
DEFINITION PREDICTED: Glycine max NADP-dependent glyceraldehyde-3-
phosphate
dehydrogenase (LOC100789108), mRNA.
ACCESSION XM_003550161
VERSION XM_003550161.4
DBLINK BioProject: PRJNA48389
KEYWORDS RefSeq.
SOURCE Glycine max (soybean)

3BIOINFORMATICS
ORGANISM Glycine max
Eukaryota; Viridiplantae; Streptophyta; Embryophyta;
Tracheophyta;
Spermatophyta; Magnoliophyta; eudicotyledons; Gunneridae;
Pentapetalae; rosids; fabids; Fabales; Fabaceae;
Papilionoideae; 50
kb inversion clade; NPAAA clade; indigoferoid/millettioid
clade;
Phaseoleae; Glycine; Soja.
COMMENT MODEL REFSEQ: This record is predicted by automated
computational
analysis. This record is derived from a genomic sequence
(NC_038253.1) annotated using gene prediction method: Gnomon,
supported by EST evidence.
Also see:
Documentation of NCBI's Annotation Process
On Aug 15, 2018 this sequence version replaced XM_003550161.3.
##Genome-Annotation-Data-START##
Annotation Provider :: NCBI
Annotation Status :: Full annotation
Annotation Name :: Glycine max Annotation Release
103
Annotation Version :: 103
Annotation Pipeline :: NCBI eukaryotic genome
annotation
pipeline
Annotation Software Version :: 8.1
Annotation Method :: Best-placed RefSeq; Gnomon
Features Annotated :: Gene; mRNA; CDS; ncRNA
##Genome-Annotation-Data-END##
FEATURES Location/Qualifiers
source 1..1941
/organism="Glycine max"
/mol_type="mRNA"
/cultivar="Williams 82"
/db_xref="taxon:3847"
/chromosome="17"
/tissue_type="callus"
gene 1..1941
/gene="LOC100789108"
/note="Derived by automated computational analysis
using
gene prediction method: Gnomon. Supporting evidence
includes similarity to: 1 EST, 19 Proteins, and 100%
coverage of the annotated genomic feature by RNAseq
alignments, including 58 samples with support for all
annotated introns"
/db_xref="GeneID:100789108"
CDS 177..1667
/gene="LOC100789108"
/codon_start=1
/product="NADP-dependent glyceraldehyde-3-phosphate
dehydrogenase"
/protein_id="XP_003550209.1"
/db_xref="GeneID:100789108"
/translation="MAGSGTFAEIIDGDVFKYYAQGHWNKSSSGKFVPIINPTTRKTH
FKVQACTQKEVNRVMESAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKE
ORGANISM Glycine max
Eukaryota; Viridiplantae; Streptophyta; Embryophyta;
Tracheophyta;
Spermatophyta; Magnoliophyta; eudicotyledons; Gunneridae;
Pentapetalae; rosids; fabids; Fabales; Fabaceae;
Papilionoideae; 50
kb inversion clade; NPAAA clade; indigoferoid/millettioid
clade;
Phaseoleae; Glycine; Soja.
COMMENT MODEL REFSEQ: This record is predicted by automated
computational
analysis. This record is derived from a genomic sequence
(NC_038253.1) annotated using gene prediction method: Gnomon,
supported by EST evidence.
Also see:
Documentation of NCBI's Annotation Process
On Aug 15, 2018 this sequence version replaced XM_003550161.3.
##Genome-Annotation-Data-START##
Annotation Provider :: NCBI
Annotation Status :: Full annotation
Annotation Name :: Glycine max Annotation Release
103
Annotation Version :: 103
Annotation Pipeline :: NCBI eukaryotic genome
annotation
pipeline
Annotation Software Version :: 8.1
Annotation Method :: Best-placed RefSeq; Gnomon
Features Annotated :: Gene; mRNA; CDS; ncRNA
##Genome-Annotation-Data-END##
FEATURES Location/Qualifiers
source 1..1941
/organism="Glycine max"
/mol_type="mRNA"
/cultivar="Williams 82"
/db_xref="taxon:3847"
/chromosome="17"
/tissue_type="callus"
gene 1..1941
/gene="LOC100789108"
/note="Derived by automated computational analysis
using
gene prediction method: Gnomon. Supporting evidence
includes similarity to: 1 EST, 19 Proteins, and 100%
coverage of the annotated genomic feature by RNAseq
alignments, including 58 samples with support for all
annotated introns"
/db_xref="GeneID:100789108"
CDS 177..1667
/gene="LOC100789108"
/codon_start=1
/product="NADP-dependent glyceraldehyde-3-phosphate
dehydrogenase"
/protein_id="XP_003550209.1"
/db_xref="GeneID:100789108"
/translation="MAGSGTFAEIIDGDVFKYYAQGHWNKSSSGKFVPIINPTTRKTH
FKVQACTQKEVNRVMESAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKE
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

4BIOINFORMATICS
IAKPAKDAVTEVIRSGDLVSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLG
VVLAIPPFNYPVNLAVSKIAPALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLI
SCVTGKGSEIGDFLTMHPGVNCISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLED
ADLDLAAANIVKGGFSYSGQRCTAVKVALVMESVANTLVKRINDKIAKLTVGPPEIDS
DVTPVVTESSANFIEGLVMDAKEKGATFCQEYVREGNLIWPLLLDNVRPDMRIAWEEP
FGPVLPVIRINSVEEGIHHCNASNFGLQGCVFTRDINKAMLISDAMETGTVQINSAPA
RGPDHFPFQGLKDSGIGSQGITNSINMMTKVKTTIINLPAPSYTMG"
ORIGIN
1 gactaagcac taagtattag caacatttat agctgttcct tgaccctgtc tgaatccact
61 tcaataatca tgttatacta ataataatca ccaaccacat aatcaacatc aatatatata
121 tactcctaag ccaaagtaca aacctcaaac caaatcgaat cacttgttag ttcaacatgg
181 ctggcagtgg cacctttgca gagatcatag atggtgatgt gttcaagtac tatgcacaag
241 gccattggaa caagtcctct tctggaaaat tcgttcccat catcaatcca accactagga
301 agacccactt caaggtccaa gcttgtacac agaaagaggt taatagggtc atggaatcag
361 caaaaacagc tcaaaaatca tgggccaaga ctccactgtg gaaaagagca gagttgcttc
421 acaaggctgc agcaatactg aaggagcaca aagcacctat tgctgagtgt cttgtaaagg
481 agattgcaaa gccagctaag gatgctgtca ctgaggtgat cagatcaggg gatttggtgt
541 catattgtgc tgaggaaggg gtgagaattc ttggggaagg aaaatttctg gtctctgata
601 gttttccggg caatgaaaga accaaatact gtctcacttc caagattcca ctgggagttg
661 ttttagccat tccacctttc aactatcctg ttaatctggc tgtttcaaag attgctccag
721 cgcttattgc tggaaactcc attgttctta aaccccccac ccagggtgct gttgctgcac
781 tacatatggt gcattgcttt catttggctg gtttccccga aggccttatt agctgtgtga
841 caggaaaggg ttccgagatt ggtgattttc ttacaatgca tccgggggtg aactgcataa
901 gtttcactgg tggggacact ggaatagcaa tatcaaagaa ggccggcatg gttcctcttc
961 aaatggaatt gggtggaaaa gatgcctgca ttgttcttga agatgctgac ttggatttgg
1021 cagctgctaa cattgtaaaa ggaggctttt cctacagtgg tcaaagatgc acagctgtaa
1081 aggttgctct ggtcatggaa tcggttgcca acactcttgt aaagagaatc aatgataaaa
1141 ttgcaaagtt aactgttgga cctcctgaga ttgacagtga tgtcacacca gttgtcaccg
1201 aatcctccgc taacttcatt gaagggttgg taatggatgc taaggagaaa ggagcaacat
1261 tctgccagga atacgtaagg gagggaaatc tcatttggcc attgctgttg gataatgttc
1321 gtcccgatat gaggatagca tgggaggagc catttggacc agttttgcca gttatcagga
1381 taaattctgt tgaagaagga atccaccatt gtaatgctag caattttggc cttcagggat
1441 gtgtcttcac aagagacatt aacaaagcaa tgctgatcag tgatgcaatg gagacaggaa
1501 cagttcaaat taattcagca ccagctcgtg gacctgatca ctttcctttt cagggtctga
1561 aggatagtgg aattggttct caagggatca ccaacagtat taacatgatg acaaaggtta
1621 agactacaat aatcaacttg ccagcacctt cttacactat gggatgagaa taatgagatg
1681 tccaagaaac agtttagtca ctcaaagtag tagtaatcag catataaagt aggcttttcc
1741 gatctcccac gttacagctt agccctatag agttgagtaa gggacagtct ttgagagggg
1801 gctgcttgca tacaaaaaat atttgttgca gtgcatgtgg attaactgta ctttatgttt
1861 ctatggagaa aagggtaatc actatttgta caagtaggat atgaatcttg aataaattat
1921 ggttctccaa ctttacttga a
//
IAKPAKDAVTEVIRSGDLVSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLG
VVLAIPPFNYPVNLAVSKIAPALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLI
SCVTGKGSEIGDFLTMHPGVNCISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLED
ADLDLAAANIVKGGFSYSGQRCTAVKVALVMESVANTLVKRINDKIAKLTVGPPEIDS
DVTPVVTESSANFIEGLVMDAKEKGATFCQEYVREGNLIWPLLLDNVRPDMRIAWEEP
FGPVLPVIRINSVEEGIHHCNASNFGLQGCVFTRDINKAMLISDAMETGTVQINSAPA
RGPDHFPFQGLKDSGIGSQGITNSINMMTKVKTTIINLPAPSYTMG"
ORIGIN
1 gactaagcac taagtattag caacatttat agctgttcct tgaccctgtc tgaatccact
61 tcaataatca tgttatacta ataataatca ccaaccacat aatcaacatc aatatatata
121 tactcctaag ccaaagtaca aacctcaaac caaatcgaat cacttgttag ttcaacatgg
181 ctggcagtgg cacctttgca gagatcatag atggtgatgt gttcaagtac tatgcacaag
241 gccattggaa caagtcctct tctggaaaat tcgttcccat catcaatcca accactagga
301 agacccactt caaggtccaa gcttgtacac agaaagaggt taatagggtc atggaatcag
361 caaaaacagc tcaaaaatca tgggccaaga ctccactgtg gaaaagagca gagttgcttc
421 acaaggctgc agcaatactg aaggagcaca aagcacctat tgctgagtgt cttgtaaagg
481 agattgcaaa gccagctaag gatgctgtca ctgaggtgat cagatcaggg gatttggtgt
541 catattgtgc tgaggaaggg gtgagaattc ttggggaagg aaaatttctg gtctctgata
601 gttttccggg caatgaaaga accaaatact gtctcacttc caagattcca ctgggagttg
661 ttttagccat tccacctttc aactatcctg ttaatctggc tgtttcaaag attgctccag
721 cgcttattgc tggaaactcc attgttctta aaccccccac ccagggtgct gttgctgcac
781 tacatatggt gcattgcttt catttggctg gtttccccga aggccttatt agctgtgtga
841 caggaaaggg ttccgagatt ggtgattttc ttacaatgca tccgggggtg aactgcataa
901 gtttcactgg tggggacact ggaatagcaa tatcaaagaa ggccggcatg gttcctcttc
961 aaatggaatt gggtggaaaa gatgcctgca ttgttcttga agatgctgac ttggatttgg
1021 cagctgctaa cattgtaaaa ggaggctttt cctacagtgg tcaaagatgc acagctgtaa
1081 aggttgctct ggtcatggaa tcggttgcca acactcttgt aaagagaatc aatgataaaa
1141 ttgcaaagtt aactgttgga cctcctgaga ttgacagtga tgtcacacca gttgtcaccg
1201 aatcctccgc taacttcatt gaagggttgg taatggatgc taaggagaaa ggagcaacat
1261 tctgccagga atacgtaagg gagggaaatc tcatttggcc attgctgttg gataatgttc
1321 gtcccgatat gaggatagca tgggaggagc catttggacc agttttgcca gttatcagga
1381 taaattctgt tgaagaagga atccaccatt gtaatgctag caattttggc cttcagggat
1441 gtgtcttcac aagagacatt aacaaagcaa tgctgatcag tgatgcaatg gagacaggaa
1501 cagttcaaat taattcagca ccagctcgtg gacctgatca ctttcctttt cagggtctga
1561 aggatagtgg aattggttct caagggatca ccaacagtat taacatgatg acaaaggtta
1621 agactacaat aatcaacttg ccagcacctt cttacactat gggatgagaa taatgagatg
1681 tccaagaaac agtttagtca ctcaaagtag tagtaatcag catataaagt aggcttttcc
1741 gatctcccac gttacagctt agccctatag agttgagtaa gggacagtct ttgagagggg
1801 gctgcttgca tacaaaaaat atttgttgca gtgcatgtgg attaactgta ctttatgttt
1861 ctatggagaa aagggtaatc actatttgta caagtaggat atgaatcttg aataaattat
1921 ggttctccaa ctttacttga a
//

5BIOINFORMATICS
Answer 2
Step 3- Searching the domain
GeneBakc full view of the conserved domain sequences
misc_feature 177..1664
/gene="LOC100789108"
/note="NADP-dependent glyceraldehyde-3-phosphate
dehydrogenase; Provisional; Region: PLN00412"
/db_xref="CDD:215110"
misc_feature order(384..389,393..401,543..545,552..557,561..566,
579..593,597..599,639..641,645..647,933..935,948..950,
969..971,1452..1454,1458..1466,1470..1475,1479..1484,
1488..1490,1494..1511,1515..1517,1536..1541,1545..1547,
1551..1553,1569..1571,1590..1595,1614..1616,1620..1637)
/gene="LOC100789108"
/note="tetrameric interface [polypeptide binding];
other
site"
/db_xref="CDD:143401"
misc_feature order(387..389,405..407,639..644)
/gene="LOC100789108"
/note="activator binding site; other site"
/db_xref="CDD:143401"
misc_feature order(669..680,750..752,756..761,849..851,864..866,
903..905,909..914,918..920,1347..1349)
/gene="LOC100789108"
/note="NADP binding site [chemical binding]; other
site"
/db_xref="CDD:143401"
misc_feature order(681..686,1065..1073,1527..1532)
/gene="LOC100789108"
/note="substrate binding site [chemical binding];
other
site"
/db_xref="CDD:143401"
misc_feature order(681..683,966..968,1059..1061,1068..1070)
/gene="LOC100789108"
/note="catalytic residues [active]"
/db_xref="CDD:143401"
Step 4 and 5- Downloading and viewing the domain containing region
Answer 2
Step 3- Searching the domain
GeneBakc full view of the conserved domain sequences
misc_feature 177..1664
/gene="LOC100789108"
/note="NADP-dependent glyceraldehyde-3-phosphate
dehydrogenase; Provisional; Region: PLN00412"
/db_xref="CDD:215110"
misc_feature order(384..389,393..401,543..545,552..557,561..566,
579..593,597..599,639..641,645..647,933..935,948..950,
969..971,1452..1454,1458..1466,1470..1475,1479..1484,
1488..1490,1494..1511,1515..1517,1536..1541,1545..1547,
1551..1553,1569..1571,1590..1595,1614..1616,1620..1637)
/gene="LOC100789108"
/note="tetrameric interface [polypeptide binding];
other
site"
/db_xref="CDD:143401"
misc_feature order(387..389,405..407,639..644)
/gene="LOC100789108"
/note="activator binding site; other site"
/db_xref="CDD:143401"
misc_feature order(669..680,750..752,756..761,849..851,864..866,
903..905,909..914,918..920,1347..1349)
/gene="LOC100789108"
/note="NADP binding site [chemical binding]; other
site"
/db_xref="CDD:143401"
misc_feature order(681..686,1065..1073,1527..1532)
/gene="LOC100789108"
/note="substrate binding site [chemical binding];
other
site"
/db_xref="CDD:143401"
misc_feature order(681..683,966..968,1059..1061,1068..1070)
/gene="LOC100789108"
/note="catalytic residues [active]"
/db_xref="CDD:143401"
Step 4 and 5- Downloading and viewing the domain containing region

6BIOINFORMATICS
Fig 3: Graphic view of the mRNA domain
Answer 3
Step 6, 7- Running BLASTn
Fig 4: Somewhat similar sequences for Glycine max
Fig 3: Graphic view of the mRNA domain
Answer 3
Step 6, 7- Running BLASTn
Fig 4: Somewhat similar sequences for Glycine max
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

7BIOINFORMATICS
Fig 5: Picture of the first alignment by BLASTn
Answer 4
Step 8, 9- Obtaining the amino acid sequences of the first five sequences from figure 4
1.
>lcl|XM_003550161.4_prot_XP_003550209.1_1 [gene=LOC100789108]
[db_xref=GeneID:100789108] [protein=NADP-dependent glyceraldehyde-3-phosphate
dehydrogenase] [protein_id=XP_003550209.1] [location=177..1667] [gbkey=CDS]
MAGSGTFAEIIDGDVFKYYAQGHWNKSSSGKFVPIINPTTRKTHFKVQACTQKEVNR
VMESAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVTE
VIRSGDLVSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLGVVLAIPPFNYPV
NLAVSKIAPALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLISCVTGKGSEIGD
FLTMHPGVNCISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLEDADLDLAAANIV
KGGFSYSGQRCTAVKVALVMESVANTLVKRINDKIAKLTVGPPEIDSDVTPVVTESS
ANFIEGLVMDAKEKGATFCQEYVREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIR
INSVEEGIHHCNASNFGLQGCVFTRDINKAMLISDAMETGTVQINSAPARGPDHFPFQ
GLKDSGIGSQGITNSINMMTKVKTTIINLPAPSYTMG
2.
Fig 5: Picture of the first alignment by BLASTn
Answer 4
Step 8, 9- Obtaining the amino acid sequences of the first five sequences from figure 4
1.
>lcl|XM_003550161.4_prot_XP_003550209.1_1 [gene=LOC100789108]
[db_xref=GeneID:100789108] [protein=NADP-dependent glyceraldehyde-3-phosphate
dehydrogenase] [protein_id=XP_003550209.1] [location=177..1667] [gbkey=CDS]
MAGSGTFAEIIDGDVFKYYAQGHWNKSSSGKFVPIINPTTRKTHFKVQACTQKEVNR
VMESAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVTE
VIRSGDLVSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLGVVLAIPPFNYPV
NLAVSKIAPALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLISCVTGKGSEIGD
FLTMHPGVNCISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLEDADLDLAAANIV
KGGFSYSGQRCTAVKVALVMESVANTLVKRINDKIAKLTVGPPEIDSDVTPVVTESS
ANFIEGLVMDAKEKGATFCQEYVREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIR
INSVEEGIHHCNASNFGLQGCVFTRDINKAMLISDAMETGTVQINSAPARGPDHFPFQ
GLKDSGIGSQGITNSINMMTKVKTTIINLPAPSYTMG
2.

8BIOINFORMATICS
>lcl|XM_028353785.1_prot_XP_028209586.1_1 [gene=LOC114392594]
[db_xref=GeneID:114392594] [protein=NADP-dependent glyceraldehyde-3-phosphate
dehydrogenase-like] [protein_id=XP_028209586.1] [location=224..1714] [gbkey=CDS]
MAGSGTFAEIIDGDVFKYYAQGHWNKSSSGKFVPIINPTTRKTHFKVQACTQKEVNR
VMESAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVTE
VIRSGDLVSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLGVVLAIPPFNYPV
NLAVSKIAPALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLISCVTGKGSEIGD
FLTMHPGVNCISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLEDADLDLAAANIV
KGGFSYSGQRCTAVKVALVMESVANTLVKKINDKIAKLTVGPPEIDSDVTPVVTESS
ANFIEGLVMDAKEKGATFCQEYVREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIR
INSVEEGIHHCNASNFGLQGCVFTRDINKAMLISDAMETGTVQINSAPARGPDHFPFQ
GLKDSGIGSQGIANSINMMTKVKTTIINLPAPSYTMG
3.
>lcl|XM_020380848.2_prot_XP_020236437.1_1 [gene=LOC109816003]
[db_xref=GeneID:109816003] [protein=NADP-dependent glyceraldehyde-3-phosphate
dehydrogenase] [protein_id=XP_020236437.1] [location=71..1561] [gbkey=CDS]
MAGSGTFAEIIDGDVFKYYSQGQWNKSSSGKFVPIINPTTRKTHFKVQACTQEEVNR
VMESAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVTE
VVRSGDLVSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLGVVLAIPPFNYP
VNLAVSKIAPALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLISCVTGKGSEIG
DFLTMHPGVNCISFTGGDTGIAISKKAGMIPLQMELGGKDACIVLEDADLDLAAANI
VKGGFSYSGQRCTAVKVALVMESVANTLVKKINDKVAKLTVGPPENDCDVTPVVT
ESSANFIEGLVMDAKEKGATFCQEYKRDGNLIWPLLLDNVRPDMRIAWEEPFGPVLP
VIRINTVEEGIHHCNASNFGLQGCVFTRDINKAMLISDAMETGTVQINSAPARGPDHF
PFQGLKDSGIGSQGITNSIDMMTKVKTTVINLPAPSYTMG
4.
>lcl|XM_017574063.1_prot_XP_017429552.1_1 [gene=LOC108337515]
[db_xref=GeneID:108337515] [protein=NADP-dependent glyceraldehyde-3-phosphate
dehydrogenase-like] [protein_id=XP_017429552.1] [location=61..1551] [gbkey=CDS]
MAATETFGEIIDGDVFKYYAHGCWNKSSSGKSVPIINPSTRKPHFKVQACTREEVDS
VMESAKTAQKSWAKTPLWKRAELLHKAAAILKEHKASIAECLVKEIAKPAKDAVTE
VVRSGDLISYCAEEGVRILGEGKFLVSDSFPGNDRTKYCLSSKIPLGVVLAIPPFNYPV
NLAVSKIAPALISGNSIVLKPPTQGAVAALHMVHCFHLAGFPQGLISCVTGKGSEIGD
FLTMHPGVNCISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLEDADLDLAAANM
VKGAFSYSGQRCTAVKVVLVMESVANTLVKKINDKVAKLTVGPPENDCDVTPVVT
ESSANFIEGLVMDAKEKGATFCQEYRREGNLIWPLLLDNVRPDMRIAWEEPFGPVLP
VIRINSVEEGIHHCNASNFGLQGCVFTKDINKAMLISDAMETGTVQINSAPSRGPDHF
PFQGLKDSGIGSQGITNSINMMTKVKTTVINLPAASYTMG
5.
>lcl|XM_007161077.1_prot_XP_007161139.1_1 [locus_tag=PHAVU_001G045700g]
[db_xref=Phytozome:Phvul.001G045700.1.p] [protein=hypothetical protein]
[protein_id=XP_007161139.1] [location=167..1657] [gbkey=CDS]
>lcl|XM_028353785.1_prot_XP_028209586.1_1 [gene=LOC114392594]
[db_xref=GeneID:114392594] [protein=NADP-dependent glyceraldehyde-3-phosphate
dehydrogenase-like] [protein_id=XP_028209586.1] [location=224..1714] [gbkey=CDS]
MAGSGTFAEIIDGDVFKYYAQGHWNKSSSGKFVPIINPTTRKTHFKVQACTQKEVNR
VMESAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVTE
VIRSGDLVSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLGVVLAIPPFNYPV
NLAVSKIAPALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLISCVTGKGSEIGD
FLTMHPGVNCISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLEDADLDLAAANIV
KGGFSYSGQRCTAVKVALVMESVANTLVKKINDKIAKLTVGPPEIDSDVTPVVTESS
ANFIEGLVMDAKEKGATFCQEYVREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIR
INSVEEGIHHCNASNFGLQGCVFTRDINKAMLISDAMETGTVQINSAPARGPDHFPFQ
GLKDSGIGSQGIANSINMMTKVKTTIINLPAPSYTMG
3.
>lcl|XM_020380848.2_prot_XP_020236437.1_1 [gene=LOC109816003]
[db_xref=GeneID:109816003] [protein=NADP-dependent glyceraldehyde-3-phosphate
dehydrogenase] [protein_id=XP_020236437.1] [location=71..1561] [gbkey=CDS]
MAGSGTFAEIIDGDVFKYYSQGQWNKSSSGKFVPIINPTTRKTHFKVQACTQEEVNR
VMESAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVTE
VVRSGDLVSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLGVVLAIPPFNYP
VNLAVSKIAPALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLISCVTGKGSEIG
DFLTMHPGVNCISFTGGDTGIAISKKAGMIPLQMELGGKDACIVLEDADLDLAAANI
VKGGFSYSGQRCTAVKVALVMESVANTLVKKINDKVAKLTVGPPENDCDVTPVVT
ESSANFIEGLVMDAKEKGATFCQEYKRDGNLIWPLLLDNVRPDMRIAWEEPFGPVLP
VIRINTVEEGIHHCNASNFGLQGCVFTRDINKAMLISDAMETGTVQINSAPARGPDHF
PFQGLKDSGIGSQGITNSIDMMTKVKTTVINLPAPSYTMG
4.
>lcl|XM_017574063.1_prot_XP_017429552.1_1 [gene=LOC108337515]
[db_xref=GeneID:108337515] [protein=NADP-dependent glyceraldehyde-3-phosphate
dehydrogenase-like] [protein_id=XP_017429552.1] [location=61..1551] [gbkey=CDS]
MAATETFGEIIDGDVFKYYAHGCWNKSSSGKSVPIINPSTRKPHFKVQACTREEVDS
VMESAKTAQKSWAKTPLWKRAELLHKAAAILKEHKASIAECLVKEIAKPAKDAVTE
VVRSGDLISYCAEEGVRILGEGKFLVSDSFPGNDRTKYCLSSKIPLGVVLAIPPFNYPV
NLAVSKIAPALISGNSIVLKPPTQGAVAALHMVHCFHLAGFPQGLISCVTGKGSEIGD
FLTMHPGVNCISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLEDADLDLAAANM
VKGAFSYSGQRCTAVKVVLVMESVANTLVKKINDKVAKLTVGPPENDCDVTPVVT
ESSANFIEGLVMDAKEKGATFCQEYRREGNLIWPLLLDNVRPDMRIAWEEPFGPVLP
VIRINSVEEGIHHCNASNFGLQGCVFTKDINKAMLISDAMETGTVQINSAPSRGPDHF
PFQGLKDSGIGSQGITNSINMMTKVKTTVINLPAASYTMG
5.
>lcl|XM_007161077.1_prot_XP_007161139.1_1 [locus_tag=PHAVU_001G045700g]
[db_xref=Phytozome:Phvul.001G045700.1.p] [protein=hypothetical protein]
[protein_id=XP_007161139.1] [location=167..1657] [gbkey=CDS]

9BIOINFORMATICS
MAGTGTFGEIIDGDVFKYYAQGHWNKSSSGKSVPIINPSTRKTQFKVQACTQEEVNR
VMESAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVSE
VVRSGDLISYCAEEGVRILGEGKLLVSDSFPGNDRTKCCLSSKIPLGVVLAIPPFNYPV
NLAVSKIAPALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPKDLISCVTGKGSEIGD
FLTMHSGVNCISFTGGDTGIAISKKAGMIPLQMELGGKDACIVLEDADLDLAAANMV
KGGFSYSGQRCTAVKVVLVIESVANTLVKKINDKVAKLTVGPPEKDSDITPVVTESS
ANFIEGLVKDAKEKGATFCQEYKREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIRI
KSVDEGINHCNASNFGLQGCVFTRDINKAMLISDAMETGTVQINSAPSRGPDHFPFQ
GLKDSGIGSQGITNSINMMTKIKTTVINLPAASYTMG
Answer 5
Step 10- Multiple sequence alignment of the five selected proteins
LUSTAL O(1.2.4) multiple sequence alignment
lcl|XM_007161077.1_prot_XP_007161139.1_1
MAGTGTFGEIIDGDVFKYYAQGHWNKSSSGKSVPIINPSTRKTQFKVQACTQEEVNRVME 60
lcl|XM_017574063.1_prot_XP_017429552.1_1
MAATETFGEIIDGDVFKYYAHGCWNKSSSGKSVPIINPSTRKPHFKVQACTREEVDSVME 60
lcl|XM_003550161.4_prot_XP_003550209.1_1
MAGSGTFAEIIDGDVFKYYAQGHWNKSSSGKFVPIINPTTRKTHFKVQACTQKEVNRVME 60
lcl|XM_028353785.1_prot_XP_028209586.1_1
MAGSGTFAEIIDGDVFKYYAQGHWNKSSSGKFVPIINPTTRKTHFKVQACTQKEVNRVME 60
lcl|XM_020380848.2_prot_XP_020236437.1_1
MAGSGTFAEIIDGDVFKYYSQGQWNKSSSGKFVPIINPTTRKTHFKVQACTQEEVNRVME 60
**.: **.***********::* ********
******:*** :*******::**: ***
lcl|XM_007161077.1_prot_XP_007161139.1_1
SAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVSEVVRSGDL 120
lcl|XM_017574063.1_prot_XP_017429552.1_1
SAKTAQKSWAKTPLWKRAELLHKAAAILKEHKASIAECLVKEIAKPAKDAVTEVVRSGDL 120
lcl|XM_003550161.4_prot_XP_003550209.1_1
SAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVTEVIRSGDL 120
lcl|XM_028353785.1_prot_XP_028209586.1_1
SAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVTEVIRSGDL 120
lcl|XM_020380848.2_prot_XP_020236437.1_1
SAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVTEVVRSGDL 120
*********************************
*****************:**:*****
lcl|XM_007161077.1_prot_XP_007161139.1_1
ISYCAEEGVRILGEGKLLVSDSFPGNDRTKCCLSSKIPLGVVLAIPPFNYPVNLAVSKIA 180
lcl|XM_017574063.1_prot_XP_017429552.1_1
ISYCAEEGVRILGEGKFLVSDSFPGNDRTKYCLSSKIPLGVVLAIPPFNYPVNLAVSKIA 180
lcl|XM_003550161.4_prot_XP_003550209.1_1
VSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLGVVLAIPPFNYPVNLAVSKIA 180
lcl|XM_028353785.1_prot_XP_028209586.1_1
VSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLGVVLAIPPFNYPVNLAVSKIA 180
lcl|XM_020380848.2_prot_XP_020236437.1_1
VSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLGVVLAIPPFNYPVNLAVSKIA 180
:***************:*********:***
**:**************************
lcl|XM_007161077.1_prot_XP_007161139.1_1
PALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPKDLISCVTGKGSEIGDFLTMHSGVNC 240
lcl|XM_017574063.1_prot_XP_017429552.1_1
PALISGNSIVLKPPTQGAVAALHMVHCFHLAGFPQGLISCVTGKGSEIGDFLTMHPGVNC 240
lcl|XM_003550161.4_prot_XP_003550209.1_1
PALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLISCVTGKGSEIGDFLTMHPGVNC 240
MAGTGTFGEIIDGDVFKYYAQGHWNKSSSGKSVPIINPSTRKTQFKVQACTQEEVNR
VMESAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVSE
VVRSGDLISYCAEEGVRILGEGKLLVSDSFPGNDRTKCCLSSKIPLGVVLAIPPFNYPV
NLAVSKIAPALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPKDLISCVTGKGSEIGD
FLTMHSGVNCISFTGGDTGIAISKKAGMIPLQMELGGKDACIVLEDADLDLAAANMV
KGGFSYSGQRCTAVKVVLVIESVANTLVKKINDKVAKLTVGPPEKDSDITPVVTESS
ANFIEGLVKDAKEKGATFCQEYKREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIRI
KSVDEGINHCNASNFGLQGCVFTRDINKAMLISDAMETGTVQINSAPSRGPDHFPFQ
GLKDSGIGSQGITNSINMMTKIKTTVINLPAASYTMG
Answer 5
Step 10- Multiple sequence alignment of the five selected proteins
LUSTAL O(1.2.4) multiple sequence alignment
lcl|XM_007161077.1_prot_XP_007161139.1_1
MAGTGTFGEIIDGDVFKYYAQGHWNKSSSGKSVPIINPSTRKTQFKVQACTQEEVNRVME 60
lcl|XM_017574063.1_prot_XP_017429552.1_1
MAATETFGEIIDGDVFKYYAHGCWNKSSSGKSVPIINPSTRKPHFKVQACTREEVDSVME 60
lcl|XM_003550161.4_prot_XP_003550209.1_1
MAGSGTFAEIIDGDVFKYYAQGHWNKSSSGKFVPIINPTTRKTHFKVQACTQKEVNRVME 60
lcl|XM_028353785.1_prot_XP_028209586.1_1
MAGSGTFAEIIDGDVFKYYAQGHWNKSSSGKFVPIINPTTRKTHFKVQACTQKEVNRVME 60
lcl|XM_020380848.2_prot_XP_020236437.1_1
MAGSGTFAEIIDGDVFKYYSQGQWNKSSSGKFVPIINPTTRKTHFKVQACTQEEVNRVME 60
**.: **.***********::* ********
******:*** :*******::**: ***
lcl|XM_007161077.1_prot_XP_007161139.1_1
SAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVSEVVRSGDL 120
lcl|XM_017574063.1_prot_XP_017429552.1_1
SAKTAQKSWAKTPLWKRAELLHKAAAILKEHKASIAECLVKEIAKPAKDAVTEVVRSGDL 120
lcl|XM_003550161.4_prot_XP_003550209.1_1
SAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVTEVIRSGDL 120
lcl|XM_028353785.1_prot_XP_028209586.1_1
SAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVTEVIRSGDL 120
lcl|XM_020380848.2_prot_XP_020236437.1_1
SAKTAQKSWAKTPLWKRAELLHKAAAILKEHKAPIAECLVKEIAKPAKDAVTEVVRSGDL 120
*********************************
*****************:**:*****
lcl|XM_007161077.1_prot_XP_007161139.1_1
ISYCAEEGVRILGEGKLLVSDSFPGNDRTKCCLSSKIPLGVVLAIPPFNYPVNLAVSKIA 180
lcl|XM_017574063.1_prot_XP_017429552.1_1
ISYCAEEGVRILGEGKFLVSDSFPGNDRTKYCLSSKIPLGVVLAIPPFNYPVNLAVSKIA 180
lcl|XM_003550161.4_prot_XP_003550209.1_1
VSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLGVVLAIPPFNYPVNLAVSKIA 180
lcl|XM_028353785.1_prot_XP_028209586.1_1
VSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLGVVLAIPPFNYPVNLAVSKIA 180
lcl|XM_020380848.2_prot_XP_020236437.1_1
VSYCAEEGVRILGEGKFLVSDSFPGNERTKYCLTSKIPLGVVLAIPPFNYPVNLAVSKIA 180
:***************:*********:***
**:**************************
lcl|XM_007161077.1_prot_XP_007161139.1_1
PALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPKDLISCVTGKGSEIGDFLTMHSGVNC 240
lcl|XM_017574063.1_prot_XP_017429552.1_1
PALISGNSIVLKPPTQGAVAALHMVHCFHLAGFPQGLISCVTGKGSEIGDFLTMHPGVNC 240
lcl|XM_003550161.4_prot_XP_003550209.1_1
PALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLISCVTGKGSEIGDFLTMHPGVNC 240
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

10BIOINFORMATICS
lcl|XM_028353785.1_prot_XP_028209586.1_1
PALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLISCVTGKGSEIGDFLTMHPGVNC 240
lcl|XM_020380848.2_prot_XP_020236437.1_1
PALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLISCVTGKGSEIGDFLTMHPGVNC 240
****:*****************************:.******************* ****
lcl|XM_007161077.1_prot_XP_007161139.1_1
ISFTGGDTGIAISKKAGMIPLQMELGGKDACIVLEDADLDLAAANMVKGGFSYSGQRCTA 300
lcl|XM_017574063.1_prot_XP_017429552.1_1
ISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLEDADLDLAAANMVKGAFSYSGQRCTA 300
lcl|XM_003550161.4_prot_XP_003550209.1_1
ISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLEDADLDLAAANIVKGGFSYSGQRCTA 300
lcl|XM_028353785.1_prot_XP_028209586.1_1
ISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLEDADLDLAAANIVKGGFSYSGQRCTA 300
lcl|XM_020380848.2_prot_XP_020236437.1_1
ISFTGGDTGIAISKKAGMIPLQMELGGKDACIVLEDADLDLAAANIVKGGFSYSGQRCTA 300
******************:**************************:***.**********
lcl|XM_007161077.1_prot_XP_007161139.1_1
VKVVLVIESVANTLVKKINDKVAKLTVGPPEKDSDITPVVTESSANFIEGLVKDAKEKGA 360
lcl|XM_017574063.1_prot_XP_017429552.1_1
VKVVLVMESVANTLVKKINDKVAKLTVGPPENDCDVTPVVTESSANFIEGLVMDAKEKGA 360
lcl|XM_003550161.4_prot_XP_003550209.1_1
VKVALVMESVANTLVKRINDKIAKLTVGPPEIDSDVTPVVTESSANFIEGLVMDAKEKGA 360
lcl|XM_028353785.1_prot_XP_028209586.1_1
VKVALVMESVANTLVKKINDKIAKLTVGPPEIDSDVTPVVTESSANFIEGLVMDAKEKGA 360
lcl|XM_020380848.2_prot_XP_020236437.1_1
VKVALVMESVANTLVKKINDKVAKLTVGPPENDCDVTPVVTESSANFIEGLVMDAKEKGA 360
***.**:*********:****:*********
*.*:**************** *******
lcl|XM_007161077.1_prot_XP_007161139.1_1
TFCQEYKREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIRIKSVDEGINHCNASNFGLQ 420
lcl|XM_017574063.1_prot_XP_017429552.1_1
TFCQEYRREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIRINSVEEGIHHCNASNFGLQ 420
lcl|XM_003550161.4_prot_XP_003550209.1_1
TFCQEYVREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIRINSVEEGIHHCNASNFGLQ 420
lcl|XM_028353785.1_prot_XP_028209586.1_1
TFCQEYVREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIRINSVEEGIHHCNASNFGLQ 420
lcl|XM_020380848.2_prot_XP_020236437.1_1
TFCQEYKRDGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIRINTVEEGIHHCNASNFGLQ 420
******
*:*********************************::*:***:**********
lcl|XM_007161077.1_prot_XP_007161139.1_1
GCVFTRDINKAMLISDAMETGTVQINSAPSRGPDHFPFQGLKDSGIGSQGITNSINMMTK 480
lcl|XM_017574063.1_prot_XP_017429552.1_1
GCVFTKDINKAMLISDAMETGTVQINSAPSRGPDHFPFQGLKDSGIGSQGITNSINMMTK 480
lcl|XM_003550161.4_prot_XP_003550209.1_1
GCVFTRDINKAMLISDAMETGTVQINSAPARGPDHFPFQGLKDSGIGSQGITNSINMMTK 480
lcl|XM_028353785.1_prot_XP_028209586.1_1
GCVFTRDINKAMLISDAMETGTVQINSAPARGPDHFPFQGLKDSGIGSQGIANSINMMTK 480
lcl|XM_020380848.2_prot_XP_020236437.1_1
GCVFTRDINKAMLISDAMETGTVQINSAPARGPDHFPFQGLKDSGIGSQGITNSIDMMTK 480
*****:***********************:*********************:***:****
lcl|XM_007161077.1_prot_XP_007161139.1_1 IKTTVINLPAASYTMG 496
lcl|XM_017574063.1_prot_XP_017429552.1_1 VKTTVINLPAASYTMG 496
lcl|XM_003550161.4_prot_XP_003550209.1_1 VKTTIINLPAPSYTMG 496
lcl|XM_028353785.1_prot_XP_028209586.1_1 VKTTIINLPAPSYTMG 496
lcl|XM_020380848.2_prot_XP_020236437.1_1 VKTTVINLPAPSYTMG 496
:***:***** *****
lcl|XM_028353785.1_prot_XP_028209586.1_1
PALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLISCVTGKGSEIGDFLTMHPGVNC 240
lcl|XM_020380848.2_prot_XP_020236437.1_1
PALIAGNSIVLKPPTQGAVAALHMVHCFHLAGFPEGLISCVTGKGSEIGDFLTMHPGVNC 240
****:*****************************:.******************* ****
lcl|XM_007161077.1_prot_XP_007161139.1_1
ISFTGGDTGIAISKKAGMIPLQMELGGKDACIVLEDADLDLAAANMVKGGFSYSGQRCTA 300
lcl|XM_017574063.1_prot_XP_017429552.1_1
ISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLEDADLDLAAANMVKGAFSYSGQRCTA 300
lcl|XM_003550161.4_prot_XP_003550209.1_1
ISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLEDADLDLAAANIVKGGFSYSGQRCTA 300
lcl|XM_028353785.1_prot_XP_028209586.1_1
ISFTGGDTGIAISKKAGMVPLQMELGGKDACIVLEDADLDLAAANIVKGGFSYSGQRCTA 300
lcl|XM_020380848.2_prot_XP_020236437.1_1
ISFTGGDTGIAISKKAGMIPLQMELGGKDACIVLEDADLDLAAANIVKGGFSYSGQRCTA 300
******************:**************************:***.**********
lcl|XM_007161077.1_prot_XP_007161139.1_1
VKVVLVIESVANTLVKKINDKVAKLTVGPPEKDSDITPVVTESSANFIEGLVKDAKEKGA 360
lcl|XM_017574063.1_prot_XP_017429552.1_1
VKVVLVMESVANTLVKKINDKVAKLTVGPPENDCDVTPVVTESSANFIEGLVMDAKEKGA 360
lcl|XM_003550161.4_prot_XP_003550209.1_1
VKVALVMESVANTLVKRINDKIAKLTVGPPEIDSDVTPVVTESSANFIEGLVMDAKEKGA 360
lcl|XM_028353785.1_prot_XP_028209586.1_1
VKVALVMESVANTLVKKINDKIAKLTVGPPEIDSDVTPVVTESSANFIEGLVMDAKEKGA 360
lcl|XM_020380848.2_prot_XP_020236437.1_1
VKVALVMESVANTLVKKINDKVAKLTVGPPENDCDVTPVVTESSANFIEGLVMDAKEKGA 360
***.**:*********:****:*********
*.*:**************** *******
lcl|XM_007161077.1_prot_XP_007161139.1_1
TFCQEYKREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIRIKSVDEGINHCNASNFGLQ 420
lcl|XM_017574063.1_prot_XP_017429552.1_1
TFCQEYRREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIRINSVEEGIHHCNASNFGLQ 420
lcl|XM_003550161.4_prot_XP_003550209.1_1
TFCQEYVREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIRINSVEEGIHHCNASNFGLQ 420
lcl|XM_028353785.1_prot_XP_028209586.1_1
TFCQEYVREGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIRINSVEEGIHHCNASNFGLQ 420
lcl|XM_020380848.2_prot_XP_020236437.1_1
TFCQEYKRDGNLIWPLLLDNVRPDMRIAWEEPFGPVLPVIRINTVEEGIHHCNASNFGLQ 420
******
*:*********************************::*:***:**********
lcl|XM_007161077.1_prot_XP_007161139.1_1
GCVFTRDINKAMLISDAMETGTVQINSAPSRGPDHFPFQGLKDSGIGSQGITNSINMMTK 480
lcl|XM_017574063.1_prot_XP_017429552.1_1
GCVFTKDINKAMLISDAMETGTVQINSAPSRGPDHFPFQGLKDSGIGSQGITNSINMMTK 480
lcl|XM_003550161.4_prot_XP_003550209.1_1
GCVFTRDINKAMLISDAMETGTVQINSAPARGPDHFPFQGLKDSGIGSQGITNSINMMTK 480
lcl|XM_028353785.1_prot_XP_028209586.1_1
GCVFTRDINKAMLISDAMETGTVQINSAPARGPDHFPFQGLKDSGIGSQGIANSINMMTK 480
lcl|XM_020380848.2_prot_XP_020236437.1_1
GCVFTRDINKAMLISDAMETGTVQINSAPARGPDHFPFQGLKDSGIGSQGITNSIDMMTK 480
*****:***********************:*********************:***:****
lcl|XM_007161077.1_prot_XP_007161139.1_1 IKTTVINLPAASYTMG 496
lcl|XM_017574063.1_prot_XP_017429552.1_1 VKTTVINLPAASYTMG 496
lcl|XM_003550161.4_prot_XP_003550209.1_1 VKTTIINLPAPSYTMG 496
lcl|XM_028353785.1_prot_XP_028209586.1_1 VKTTIINLPAPSYTMG 496
lcl|XM_020380848.2_prot_XP_020236437.1_1 VKTTVINLPAPSYTMG 496
:***:***** *****

11BIOINFORMATICS
Answer 6
Step 11- Phylogenetic tree production
Fig 6: Phylogenetic tree
Answer 6
Step 11- Phylogenetic tree production
Fig 6: Phylogenetic tree

12BIOINFORMATICS
Tools used:
Clustal W
Clustal omega
Databases:
NCBI, UniProt
Tools used:
Clustal W
Clustal omega
Databases:
NCBI, UniProt
1 out of 13
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
© 2024 | Zucol Services PVT LTD | All rights reserved.