Applications of CRISPR-Cas Technology in S. thermophilus Bacterial Adaptive Immunity
VerifiedAdded on 2023/04/25
|21
|7383
|400
AI Summary
This research focuses on the application of CRISPR-Cas technology in S. thermophilus bacterial adaptive immunity. It discusses the use of S. Thermophilus with the help of CRISPR CAS system, gene editing, and factors affecting gene editing.
Contribute Materials
Your contribution can guide someone’s learning journey. Share your
documents today.

Applications of CRISPR-Cas Technology in
S. thermophilus Bacterial Adaptive
Immunity
S. thermophilus Bacterial Adaptive
Immunity
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

Application of CRISPR-Cas Technology
Lay out Summary
CRISPR stands for clustered regularly interspaced short palindromic repeats while Cas stands for
CRISPR-associated protein. CRISPR is a group of DNA chain that can be established within the
genes of prokaryotic creatures such as archaea and bacteria. The sequences involved are imitated
form DNA portions form viruses due to which prokaryotes are infected before and are used to
expose and shatter DNA from alike viruses all along consequent infections.
Analyzation of genes with the help of CRISPR CAS
CRISPR CAS is used to analyze genes with creatures.
Cas(termed as CRISPR-associated protein) is a type of enzyme CRISPR progressions as a
mentor cleave and recognize definite strings of DNA equivalent to CRISPR progressions. Cas
combines with CRISPR sequences in order to form the origin of a mechanism that is called
CRISPR-Cas9.
Together CRISPR-Cas constitutes a system termed as a prokaryotic immune system which
deliberates opposition to external genetic elements including those existing within phages and
plasmids which supports a system of acquired immunity.
Application in various fields
CRISPR is a very huge and widely used technology that has its application in various fields.
More than 1000 research paper had been published mentioning CRISPR. It is used to activate
static or unused genes of the human body, used to analyze ‘candida albicans’, to transform yeast,
etc.
Theme
The theme of the above research is that it focuses on putting the light on those areas where there
is a huge application of CRISPR CAS system, whether it is used in transforming yeasts or
altering genes.
2 | P a g e
Lay out Summary
CRISPR stands for clustered regularly interspaced short palindromic repeats while Cas stands for
CRISPR-associated protein. CRISPR is a group of DNA chain that can be established within the
genes of prokaryotic creatures such as archaea and bacteria. The sequences involved are imitated
form DNA portions form viruses due to which prokaryotes are infected before and are used to
expose and shatter DNA from alike viruses all along consequent infections.
Analyzation of genes with the help of CRISPR CAS
CRISPR CAS is used to analyze genes with creatures.
Cas(termed as CRISPR-associated protein) is a type of enzyme CRISPR progressions as a
mentor cleave and recognize definite strings of DNA equivalent to CRISPR progressions. Cas
combines with CRISPR sequences in order to form the origin of a mechanism that is called
CRISPR-Cas9.
Together CRISPR-Cas constitutes a system termed as a prokaryotic immune system which
deliberates opposition to external genetic elements including those existing within phages and
plasmids which supports a system of acquired immunity.
Application in various fields
CRISPR is a very huge and widely used technology that has its application in various fields.
More than 1000 research paper had been published mentioning CRISPR. It is used to activate
static or unused genes of the human body, used to analyze ‘candida albicans’, to transform yeast,
etc.
Theme
The theme of the above research is that it focuses on putting the light on those areas where there
is a huge application of CRISPR CAS system, whether it is used in transforming yeasts or
altering genes.
2 | P a g e
Aims and objectives
● To analyze various types of gene editing
● To study about an architecture of S. thermophilus CRISPR 3 and S. pyogenes type II CRISPR-
Cas loci
● To examine a novel Streptococcus thermophilus locus which is termed as CRISPR 3
● To provide a new method in relation to CRISPR CAS technology
Timescale
Main Activities/ Stages
Week 1 Week 2 Week 3 Week 4 Week 5 Week 6
Topic selection and its scope
Secondary data sources
identification
Research proposal preparation
Preparation of literature review
Description of research methodology
Analysing data
Findings comparison
Conclusion and recommendations
Finalising and submission
S. Thermophilus under CRISPR CAS
CRISPR-Cas locus provides a particular study about the immunity against external components
which are more variable amidst various prokaryotes. The main purpose of the above research is
to focus on terminating the editing of genome protocols found in Streptococcus thermophilus. At
first, those protocols and conditions are introduced that play a role in genome editing. Moreover,
categorization and variety related investigation S. thermophilus CRISPR-Cas help in
3 | P a g e
● To analyze various types of gene editing
● To study about an architecture of S. thermophilus CRISPR 3 and S. pyogenes type II CRISPR-
Cas loci
● To examine a novel Streptococcus thermophilus locus which is termed as CRISPR 3
● To provide a new method in relation to CRISPR CAS technology
Timescale
Main Activities/ Stages
Week 1 Week 2 Week 3 Week 4 Week 5 Week 6
Topic selection and its scope
Secondary data sources
identification
Research proposal preparation
Preparation of literature review
Description of research methodology
Analysing data
Findings comparison
Conclusion and recommendations
Finalising and submission
S. Thermophilus under CRISPR CAS
CRISPR-Cas locus provides a particular study about the immunity against external components
which are more variable amidst various prokaryotes. The main purpose of the above research is
to focus on terminating the editing of genome protocols found in Streptococcus thermophilus. At
first, those protocols and conditions are introduced that play a role in genome editing. Moreover,
categorization and variety related investigation S. thermophilus CRISPR-Cas help in
3 | P a g e

understanding the connection among Streptococcus. Also, its ability to differentiate external
segments and spacer source investigation also signifies a new way of phase opposition.
The system of CRISPR-Cas was brought into being in thermophilic archaea for the very first
time and was utilized to combat external infections related to DNA (Garneau et al., 2010). It
constitutes Cas and CRISPR protein, the anterior is a part of a common group of short-layoff
palindromes which consist of various duplicative progressions and the following one is a type of
protein which is correlated with helicase activity, nuclease, and CRISPRs genes (Chylinski et al.,
2014). The structure of the system of CRISPR-Cas is more alike RNA interference. There are a
total of three stages included in its immune system namely interference, adaption and expression
(Choi and Lee, 2016). Moreover, the new system of classification that mainly includes 3 types
was suggested by Makarova et al. (2011).
Background
CRISPR CAS is a widely used/applied technology which helps in many ways to cure some
serious diseases including cancer, or altering genes. The above research focuses on providing a
context to the use of S. Thermophilus with the help of CRISPR CAS system. CRISPR CAS
changed the history of revolution after its foundation and now it plays a major role in genome
editing.
Themes
Use of S. Thermophilus
A very important and most beneficial lactic acid bacteria termed as Streptococcus thermophilus
is broadly adopted in the generation of dairy products. Additionally, its use in the field of
CRISPR Cas system appears to be one more popular area of research. After the continuing
process of evolution and immunity, S. thermophilus CRISPR Cas loci provides abundant
varieties. In accordance with dissimilar genetic location, duplicative progressions, and Cas
genes, they are divided into 4 major types- CRISPR 1, CRISPR 2, CRISPR 3, and CRISPR 4.
All of these are further enclosed in 3 types of CRISPR Cas system. The sequences of the
CRISPR Cas system differ from part to part due to the reason that there is a difference in the
amount of space.
4 | P a g e
segments and spacer source investigation also signifies a new way of phase opposition.
The system of CRISPR-Cas was brought into being in thermophilic archaea for the very first
time and was utilized to combat external infections related to DNA (Garneau et al., 2010). It
constitutes Cas and CRISPR protein, the anterior is a part of a common group of short-layoff
palindromes which consist of various duplicative progressions and the following one is a type of
protein which is correlated with helicase activity, nuclease, and CRISPRs genes (Chylinski et al.,
2014). The structure of the system of CRISPR-Cas is more alike RNA interference. There are a
total of three stages included in its immune system namely interference, adaption and expression
(Choi and Lee, 2016). Moreover, the new system of classification that mainly includes 3 types
was suggested by Makarova et al. (2011).
Background
CRISPR CAS is a widely used/applied technology which helps in many ways to cure some
serious diseases including cancer, or altering genes. The above research focuses on providing a
context to the use of S. Thermophilus with the help of CRISPR CAS system. CRISPR CAS
changed the history of revolution after its foundation and now it plays a major role in genome
editing.
Themes
Use of S. Thermophilus
A very important and most beneficial lactic acid bacteria termed as Streptococcus thermophilus
is broadly adopted in the generation of dairy products. Additionally, its use in the field of
CRISPR Cas system appears to be one more popular area of research. After the continuing
process of evolution and immunity, S. thermophilus CRISPR Cas loci provides abundant
varieties. In accordance with dissimilar genetic location, duplicative progressions, and Cas
genes, they are divided into 4 major types- CRISPR 1, CRISPR 2, CRISPR 3, and CRISPR 4.
All of these are further enclosed in 3 types of CRISPR Cas system. The sequences of the
CRISPR Cas system differ from part to part due to the reason that there is a difference in the
amount of space.
4 | P a g e
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

Application of S. thermophilus CRISPR Cas system in the field of gene editing
When the CRISPR system was compared with intensely small recombineering adeptness which
is assumed to be 10-4 to 10-6 (Sharan et al., 2009) and tedious expansion of classical methods of
homologous recombination, it could undoubtedly clarify the mutant architecture due to the
reason that it has high efficiency of editing (nearly about 50 to 100%) and chances to elite dual-
crossover cases in single step (Chen et al., 2017).
The start of the CRISPR Cas system is from progression definite identification and object DNA
loci cleavage. The termination of the system is with permanent mutations over DNA restoration
system (Selle and Barrangou, 2015). Currently, the system of CRISPR-Cas associated with S.
pyogenes (SpCas9) is broadly enforced as an appliance in gene alteration. Nevertheless, weak
points of the above system are recorded as its additional studies and considerable application
(Hsu et al., 2013). Due to the same reason, the focus of researchers is more on the S.
thermophilus CRISPR Cas system that provides inferior targeting capability and lesser Cas
portion (Xu et al., 2015). In the opion of Müller et al., (2016) the main process of this system
contains the building of working vector that contains the progression of StCas9 system and
interpretation analysis in the host cell.
Basis of similarities between StCas9 and SpCas9
Various researches propose that the cleavage activities shown by StCas9 system are very similar
to that of SpCas9 system and workability of gene editing in plants, human and microorganisms
(Xu et al., 2015; Müller et al., 2016; Steinert et al., 2016). The Cas9 protein is one of the
essential parts of the StCas system. As the director of probing action, the compositions of Cas9
include a recognition(REC) lobe and a nuclease (NUC) (Anders et al., 2014; Nishimasu et al.,
2014). Before, the PAM-definite probing of destination DNA dual strand is mainly due to RuvC,
NUC alike nuclease and PI domains. And the further is accountable for identification of sgRNA
destination DNA complex (Chen et al., 2017). St1Cas9 and St3Cas9 are included in the StCas9
system that further present image of S. thermophilus CRISPR Cas system in CRISPR 1 and
CRISPR 3 loci respectively. St1Cas9 addresses 5′-NNAGAAW while St3Cas9 addresses 5′-
5 | P a g e
When the CRISPR system was compared with intensely small recombineering adeptness which
is assumed to be 10-4 to 10-6 (Sharan et al., 2009) and tedious expansion of classical methods of
homologous recombination, it could undoubtedly clarify the mutant architecture due to the
reason that it has high efficiency of editing (nearly about 50 to 100%) and chances to elite dual-
crossover cases in single step (Chen et al., 2017).
The start of the CRISPR Cas system is from progression definite identification and object DNA
loci cleavage. The termination of the system is with permanent mutations over DNA restoration
system (Selle and Barrangou, 2015). Currently, the system of CRISPR-Cas associated with S.
pyogenes (SpCas9) is broadly enforced as an appliance in gene alteration. Nevertheless, weak
points of the above system are recorded as its additional studies and considerable application
(Hsu et al., 2013). Due to the same reason, the focus of researchers is more on the S.
thermophilus CRISPR Cas system that provides inferior targeting capability and lesser Cas
portion (Xu et al., 2015). In the opion of Müller et al., (2016) the main process of this system
contains the building of working vector that contains the progression of StCas9 system and
interpretation analysis in the host cell.
Basis of similarities between StCas9 and SpCas9
Various researches propose that the cleavage activities shown by StCas9 system are very similar
to that of SpCas9 system and workability of gene editing in plants, human and microorganisms
(Xu et al., 2015; Müller et al., 2016; Steinert et al., 2016). The Cas9 protein is one of the
essential parts of the StCas system. As the director of probing action, the compositions of Cas9
include a recognition(REC) lobe and a nuclease (NUC) (Anders et al., 2014; Nishimasu et al.,
2014). Before, the PAM-definite probing of destination DNA dual strand is mainly due to RuvC,
NUC alike nuclease and PI domains. And the further is accountable for identification of sgRNA
destination DNA complex (Chen et al., 2017). St1Cas9 and St3Cas9 are included in the StCas9
system that further present image of S. thermophilus CRISPR Cas system in CRISPR 1 and
CRISPR 3 loci respectively. St1Cas9 addresses 5′-NNAGAAW while St3Cas9 addresses 5′-
5 | P a g e

NGGNG (Garneau et al., 2010; Magadán et al., 2012). Thus, the destination capability shown by
both the system is lightly dissimilar. The efficiency presented by St3Cas system is higher than
the efficiency presented by St1Cas system (Müller et al., 2016).
Nevertheless, both the systems can be utilized in the modification of genome in accordance with
existing researches (Xu et al., 2015; Müller et al., 2016; Steinert et al., 2016).
Factors affecting gene editing
In addition, there are various important factors that affect editing capability and activities of
StCas9 system. First of all, there is sgRNA that consist of tracrRNA (trans-activating RNA) and
crRNA (CRISPR RNA) that possesses its usual stem-loop architecture which can improve the
synergy among sgRNA and Cas9. But ring and stem have authority over its action. For example,
a stem-circle structure in 3′ end of tracrRNA assumes crucial jobs in action of StCas9. What's
more, its inward lumps are fundamental for the elements of StCas9 while the difference inside
circle arrangements has no impact. (Xu et al., 2015). As noted, crRNA is the structured spacer
following up on the target DNA strand to give explicit focusing on. tracrRNA is in charge of
Cas9 blend and its 3′ locale is fundamental to StCas9 action (Wiedenheft et al., 2012; Xu et al.,
2015). Moreover, the measurements of sgRNA and Cas9 is firmly identified with the askew
effectiveness of StCas9 framework while abbreviated tracrRNA could raise its indicating
capability. Also, PAM and its close-by bases are critical for explicit acknowledgement of target
pieces. Particularly, G in PAM a respectable halfway point is fundamental for the action of
StCas9. Taking everything into account, StCas9 framework cannot exclusively be generally
utilized in the editing of genes yet in addition present increasingly dynamic application
prospects.
New method for amplification of products
Moreover, a new method termed as CRISPR array assembling method which is based on spacer
S. thermophilus has been described. Along these lines, this can be utilized as a leap forward of
helpful multiplex focusing on. First of all, tracrRNA tape, cas9 part, and CRISPR pioneer
arrangement together with CRISPR eliminator ought to be gotten by amplifying of PCR.
6 | P a g e
both the system is lightly dissimilar. The efficiency presented by St3Cas system is higher than
the efficiency presented by St1Cas system (Müller et al., 2016).
Nevertheless, both the systems can be utilized in the modification of genome in accordance with
existing researches (Xu et al., 2015; Müller et al., 2016; Steinert et al., 2016).
Factors affecting gene editing
In addition, there are various important factors that affect editing capability and activities of
StCas9 system. First of all, there is sgRNA that consist of tracrRNA (trans-activating RNA) and
crRNA (CRISPR RNA) that possesses its usual stem-loop architecture which can improve the
synergy among sgRNA and Cas9. But ring and stem have authority over its action. For example,
a stem-circle structure in 3′ end of tracrRNA assumes crucial jobs in action of StCas9. What's
more, its inward lumps are fundamental for the elements of StCas9 while the difference inside
circle arrangements has no impact. (Xu et al., 2015). As noted, crRNA is the structured spacer
following up on the target DNA strand to give explicit focusing on. tracrRNA is in charge of
Cas9 blend and its 3′ locale is fundamental to StCas9 action (Wiedenheft et al., 2012; Xu et al.,
2015). Moreover, the measurements of sgRNA and Cas9 is firmly identified with the askew
effectiveness of StCas9 framework while abbreviated tracrRNA could raise its indicating
capability. Also, PAM and its close-by bases are critical for explicit acknowledgement of target
pieces. Particularly, G in PAM a respectable halfway point is fundamental for the action of
StCas9. Taking everything into account, StCas9 framework cannot exclusively be generally
utilized in the editing of genes yet in addition present increasingly dynamic application
prospects.
New method for amplification of products
Moreover, a new method termed as CRISPR array assembling method which is based on spacer
S. thermophilus has been described. Along these lines, this can be utilized as a leap forward of
helpful multiplex focusing on. First of all, tracrRNA tape, cas9 part, and CRISPR pioneer
arrangement together with CRISPR eliminator ought to be gotten by amplifying of PCR.
6 | P a g e

After that, the products that are going to be amplified are put together in a suitable direction with
specific constraints endonucleases area. With respect to multiplex focusing on, the procedure of
DR (direct rehash)- Spacer direction is embraced rather than sgRNA direction. Legitimate
preliminaries ought to be incorporated for enhancing of intrigued grouping containing required
DR arrays and explicit spacers right off the bat. Especially, when structuring groundworks,
thought of holding relating limitation destinations is important. Through specifically toughening
oligonucleotides, the first DR exhibit can be essentially gathered into essential CRISPR vector.
After that, primers are utilized to assemble and extend consequent spacers and DR progressions
into CRISPR location to construct an artificial structure of Spacer-dr (Gou et al., 2007).
Assortments of explicit vectors going for various target genes can be remade by along these
lines. At long last, the following recombinant plasmid is moved into beneficiary cells to get
target freaks, and phenotype perception and sequencing are completed to measure its altering
productivity.
Saddling chemically dormant Cas9, whose endonucleolytic movement has been dispensed with
yet recognizable proof and restricting capacities are as yet held, together with specific
interpretation aspects forestalling or encouraging the banding of RNA polymerase (RNAP) and
its advertiser, a proficient translation guideline can be accomplished (Bikard et al., 2013).
The evolutionary relationship of S. thermophilus CRISPR Loci and their diversity
In accordance with CRISPR progression instruction of various acknowledged genome exertions
written in CRISPR directory, the category of CRISPR loci in most of the S. thermophilus strains
are of different types (more than one). The main four categories of CRISPR system can be
classified by different allocation or arrangements and DR of Cas genes. In addition, there are
various distributions of CRISPR Cas system in S. thermophilus ND07.
Universally, the dynamic idea of CRISPR loci may demonstrate important for composing and
near investigations of strains and microbial populaces. Likewise, CRISPRs give basic
experiences into the connections among prokaryotes and their surroundings, quite the
coevolution of viral and host genomes.
CRISPR4 chiefly works through antiviral guard intervened by Cascade superfamily. What's
more, its cas2 stop codon agrees with halfway grouping of the recurrent component. There is no
7 | P a g e
specific constraints endonucleases area. With respect to multiplex focusing on, the procedure of
DR (direct rehash)- Spacer direction is embraced rather than sgRNA direction. Legitimate
preliminaries ought to be incorporated for enhancing of intrigued grouping containing required
DR arrays and explicit spacers right off the bat. Especially, when structuring groundworks,
thought of holding relating limitation destinations is important. Through specifically toughening
oligonucleotides, the first DR exhibit can be essentially gathered into essential CRISPR vector.
After that, primers are utilized to assemble and extend consequent spacers and DR progressions
into CRISPR location to construct an artificial structure of Spacer-dr (Gou et al., 2007).
Assortments of explicit vectors going for various target genes can be remade by along these
lines. At long last, the following recombinant plasmid is moved into beneficiary cells to get
target freaks, and phenotype perception and sequencing are completed to measure its altering
productivity.
Saddling chemically dormant Cas9, whose endonucleolytic movement has been dispensed with
yet recognizable proof and restricting capacities are as yet held, together with specific
interpretation aspects forestalling or encouraging the banding of RNA polymerase (RNAP) and
its advertiser, a proficient translation guideline can be accomplished (Bikard et al., 2013).
The evolutionary relationship of S. thermophilus CRISPR Loci and their diversity
In accordance with CRISPR progression instruction of various acknowledged genome exertions
written in CRISPR directory, the category of CRISPR loci in most of the S. thermophilus strains
are of different types (more than one). The main four categories of CRISPR system can be
classified by different allocation or arrangements and DR of Cas genes. In addition, there are
various distributions of CRISPR Cas system in S. thermophilus ND07.
Universally, the dynamic idea of CRISPR loci may demonstrate important for composing and
near investigations of strains and microbial populaces. Likewise, CRISPRs give basic
experiences into the connections among prokaryotes and their surroundings, quite the
coevolution of viral and host genomes.
CRISPR4 chiefly works through antiviral guard intervened by Cascade superfamily. What's
more, its cas2 stop codon agrees with halfway grouping of the recurrent component. There is no
7 | P a g e
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

novel spacer expansion when remote DNA attacking, however huge up-managed articulation of
quality encoding Cascade complex can be watched, particularly cas7 (Young et al., 2012).
CRISPR 4 experiences a one of a kind interpretation process and assumes unmistakable jobs
amid phage opposition. Novel spacers are adept to assume increasingly vital jobs on opposing
phages. Of note, spacer erasure has been identified in bacteriophage-insensitive S. thermophilus
freaks (BIMs) amid phage opposition process. What's more, the erasure dependably happens at
the contiguous position with expansion, suggesting the probability that cancellation occurs by
virtue of homologous recombination between DR arrangements (Deveau et al., 2009).
The above figure represents the features of CRISPR loci
Ordinarily, bunched, routinely interspaced short palindromic rehashes (CRISPRs, white boxes)
are gone before by a pioneer arrangement (black box) which at is AT-rich however generally not
rationed. The number of rehashes can differ considerably, from at least two to a couple of
hundred. Rehash length, be that as it may, is confined to 23 to 50 nucleotides. Rehashes are
isolated by correspondingly measured, non-dreary spacers (hued boxes) that share arrangement
personality with parts of plasmids and bacteriophage genomes and determine the objectives of
CRISPR impedance. A lot of CRISPR-related (cas) qualities promptly goes before or pursues the
rehashes. These qualities are monitored, can be ordered into various families and subtypes, and
encode the protein apparatus in charge of CRISPR action.
Methodological approach
There are various methods used in the above research in order to find a relevance including the
main title of the research. A method named “array addressing “ has been used in the above
section which is based upon spacer S. Thermophilus.
8 | P a g e
quality encoding Cascade complex can be watched, particularly cas7 (Young et al., 2012).
CRISPR 4 experiences a one of a kind interpretation process and assumes unmistakable jobs
amid phage opposition. Novel spacers are adept to assume increasingly vital jobs on opposing
phages. Of note, spacer erasure has been identified in bacteriophage-insensitive S. thermophilus
freaks (BIMs) amid phage opposition process. What's more, the erasure dependably happens at
the contiguous position with expansion, suggesting the probability that cancellation occurs by
virtue of homologous recombination between DR arrangements (Deveau et al., 2009).
The above figure represents the features of CRISPR loci
Ordinarily, bunched, routinely interspaced short palindromic rehashes (CRISPRs, white boxes)
are gone before by a pioneer arrangement (black box) which at is AT-rich however generally not
rationed. The number of rehashes can differ considerably, from at least two to a couple of
hundred. Rehash length, be that as it may, is confined to 23 to 50 nucleotides. Rehashes are
isolated by correspondingly measured, non-dreary spacers (hued boxes) that share arrangement
personality with parts of plasmids and bacteriophage genomes and determine the objectives of
CRISPR impedance. A lot of CRISPR-related (cas) qualities promptly goes before or pursues the
rehashes. These qualities are monitored, can be ordered into various families and subtypes, and
encode the protein apparatus in charge of CRISPR action.
Methodological approach
There are various methods used in the above research in order to find a relevance including the
main title of the research. A method named “array addressing “ has been used in the above
section which is based upon spacer S. Thermophilus.
8 | P a g e

Moreover, the data used to conduct the research is secondary. The main advantage of using a
secondary data is that you do not have to perform a particular set of instructions again and again.
The values for them are provided in general. The experimental plan included in the above section
defines a way in which a data should be collected, analysed, used and presented.
A new method
A new method of addressing CRISPR termed as array assembling has been discussed in the
previous section. The method is based upon spacer S. thermophilus.
Thusly, this can be used as a jump forward of accommodating multiplex concentrating on.
Above all else, tracrRNA tape, cas9 part, and CRISPR pioneer game plan together with CRISPR
eliminator should be gotten by enhancing of PCR.
Purpose of the study
The main purpose and focus of this research are spread on various technologies of gene editing,
growth-related connection, and anti-phage actions of S. thermophilus.
The study provides a new method that can be further useful in the field of CRISPR CAS
technology.
Explanation of the method used
From that point forward, the items that will be enhanced are assembled an appropriate way with
explicit imperatives endonucleases region. Concerning multiplex concentrating on, the system of
DR (direct repeat)- Spacer course is grasped instead of sgRNA bearing. Real fundamentals
should be fused for improving captivated gathering containing required DR exhibits and express
spacers directly off the bat. Particularly, while organizing foundations, thought of holding
relating impediment goals is essential. Through explicitly toughening oligonucleotides, the first
DR show can be basically assembled into basic CRISPR vector. From that point forward,
preliminaries are used to amass and broaden subsequent spacers and DR movements into
9 | P a g e
secondary data is that you do not have to perform a particular set of instructions again and again.
The values for them are provided in general. The experimental plan included in the above section
defines a way in which a data should be collected, analysed, used and presented.
A new method
A new method of addressing CRISPR termed as array assembling has been discussed in the
previous section. The method is based upon spacer S. thermophilus.
Thusly, this can be used as a jump forward of accommodating multiplex concentrating on.
Above all else, tracrRNA tape, cas9 part, and CRISPR pioneer game plan together with CRISPR
eliminator should be gotten by enhancing of PCR.
Purpose of the study
The main purpose and focus of this research are spread on various technologies of gene editing,
growth-related connection, and anti-phage actions of S. thermophilus.
The study provides a new method that can be further useful in the field of CRISPR CAS
technology.
Explanation of the method used
From that point forward, the items that will be enhanced are assembled an appropriate way with
explicit imperatives endonucleases region. Concerning multiplex concentrating on, the system of
DR (direct repeat)- Spacer course is grasped instead of sgRNA bearing. Real fundamentals
should be fused for improving captivated gathering containing required DR exhibits and express
spacers directly off the bat. Particularly, while organizing foundations, thought of holding
relating impediment goals is essential. Through explicitly toughening oligonucleotides, the first
DR show can be basically assembled into basic CRISPR vector. From that point forward,
preliminaries are used to amass and broaden subsequent spacers and DR movements into
9 | P a g e

CRISPR area to develop a counterfeit structure of Spacer-dr. Combinations of express vectors
going for different target qualities can be changed by thusly. Finally, the accompanying
recombinant plasmid is moved into recipient cells to get target monstrosities, and phenotype
recognition and sequencing are finished to gauge its adjusting profitability.
Saddling artificially lethargic Cas9, whose endonucleolytic development has been shed yet
conspicuous evidence and confining limits are up 'til now held, together with explicit
understanding angles thwarting or empowering the banding of RNA polymerase (RNAP) and its
promoter, a capable interpretation rule can be cultivated.
Types of CRISPR Cas elements
There are a total of four types of CRISPR Cas elements. CRISPR 1 and CRISPR 3 are associated
with Csn type-2 A. both the CRISPR loci have an important aspect in interference and
adjustment stages. The division and mechanism of Cas progressions in CRISPR 3 and CRISPR 1
loci are comparably moderate having the extent of Cas genes in efforts of their duplicative spacer
areas. Nevertheless, the progression comparison is small and similar. It presents 33.6% identity
in Cas 1 genes and 41.3% identity in Cas 2 genes. In addition, to which, all of them exhibit the
strong capability of the combination of unique spacers. There are various experiments that
conclude the same thing.
With the help of vitro experiments, S. thermophilus can normally acquire spacers from plasmic
DNA bacteriophages into its CRISPR 3 and CRISPR 1 (Garneau et al., 2010). This appearance
can perform a better process of spacer combination as well as it contributes to a deep recognition
of external DNA. moreover, CRISPR 1 is extensively dispersed in S. thermophilus.
When it comes to CRISPR 2, it is included in the system of Csm type-3 A. it is very dissimilar in
both of the duplicating progressions and dispersion of Cas genes, having an identity of 27.78% in
the sequences of DR and Cas genes situated together with the duplicating spacer allocation. Plus,
with respect to CRISPR 2 locus, high extent of Cas however low level of rehash spacer locale
result in its bad capacity of coordinating latest spacers. Dispersion of CRISPR loci demonstrates
that CRISPR2 is most likely a bit of deteriorated part started from the gram-positive precursor
(Horvath et al., 2008).
10 | P a g e
going for different target qualities can be changed by thusly. Finally, the accompanying
recombinant plasmid is moved into recipient cells to get target monstrosities, and phenotype
recognition and sequencing are finished to gauge its adjusting profitability.
Saddling artificially lethargic Cas9, whose endonucleolytic development has been shed yet
conspicuous evidence and confining limits are up 'til now held, together with explicit
understanding angles thwarting or empowering the banding of RNA polymerase (RNAP) and its
promoter, a capable interpretation rule can be cultivated.
Types of CRISPR Cas elements
There are a total of four types of CRISPR Cas elements. CRISPR 1 and CRISPR 3 are associated
with Csn type-2 A. both the CRISPR loci have an important aspect in interference and
adjustment stages. The division and mechanism of Cas progressions in CRISPR 3 and CRISPR 1
loci are comparably moderate having the extent of Cas genes in efforts of their duplicative spacer
areas. Nevertheless, the progression comparison is small and similar. It presents 33.6% identity
in Cas 1 genes and 41.3% identity in Cas 2 genes. In addition, to which, all of them exhibit the
strong capability of the combination of unique spacers. There are various experiments that
conclude the same thing.
With the help of vitro experiments, S. thermophilus can normally acquire spacers from plasmic
DNA bacteriophages into its CRISPR 3 and CRISPR 1 (Garneau et al., 2010). This appearance
can perform a better process of spacer combination as well as it contributes to a deep recognition
of external DNA. moreover, CRISPR 1 is extensively dispersed in S. thermophilus.
When it comes to CRISPR 2, it is included in the system of Csm type-3 A. it is very dissimilar in
both of the duplicating progressions and dispersion of Cas genes, having an identity of 27.78% in
the sequences of DR and Cas genes situated together with the duplicating spacer allocation. Plus,
with respect to CRISPR 2 locus, high extent of Cas however low level of rehash spacer locale
result in its bad capacity of coordinating latest spacers. Dispersion of CRISPR loci demonstrates
that CRISPR2 is most likely a bit of deteriorated part started from the gram-positive precursor
(Horvath et al., 2008).
10 | P a g e
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

In the case of CRISPR 4, it belongs to the system of Cse type 1-E and is present in few strains
only. CRISPR 4 experiences an exclusive inscription process and performs different roles
meanwhile phage opposition. The percentage of repeating progression is 28% only diversified
Cas modules are dispersed crucially over the duplicative spacer area. There is no novel spacer
expansion when outside DNA attacking, however huge up-controlled articulation of quality
encoding Cascade complex can be watched, particularly cas7. Hence, CRISPR 4 primarily
performs over antiviral armament interposed by the Cascade family.
Conclusion of dispersion
It is concluded that the dispersion of Cas 1 and Cas 2 has concentrated authorities over
procurement of attacking progression. They are capable of forming a DNA complex termed as
Cas 1- Cas 2 protospacer when nominating a specific external DNA. Also, both the clusters
possess amid all the system of CRISPR Cas and introduce preservation that can be used in the
conditional genomic analysis. It has been determined that the new complex formed (Cas 1- Cas
2) behaves as an integrating agent novel spacer in CRISPR locus. Moreover, the latest spacers
are likely to aggregate into the end which is leading the duplicative spacer area (5’ area) although
latent spacers are suitable to be removed from the tail end of the duplicative spacer area (3’ end).
The occurrence of integration and deletion process can be simultaneous. Hence, the CRISPR
leading end displays high uncertainty while the CRISPR tail end shows high equality amid
various strains. Research determined that regularity among CRISPR and Cas protein is in or by
comparison extreme in similar type. This consideration signifies coevolution and interrelation of
mutual support among them. Also, there are critical similarities of various Cas protein amidst
different species, for instance, Cas 7 and Cas 9 all of which play an important part in protecting
external DNA. The research states that CRISPR 1 and CRISPR 4 are present in a few species
only while CRISPR is commonly found in most of the Streptococcus species.
Impact of the above system
11 | P a g e
only. CRISPR 4 experiences an exclusive inscription process and performs different roles
meanwhile phage opposition. The percentage of repeating progression is 28% only diversified
Cas modules are dispersed crucially over the duplicative spacer area. There is no novel spacer
expansion when outside DNA attacking, however huge up-controlled articulation of quality
encoding Cascade complex can be watched, particularly cas7. Hence, CRISPR 4 primarily
performs over antiviral armament interposed by the Cascade family.
Conclusion of dispersion
It is concluded that the dispersion of Cas 1 and Cas 2 has concentrated authorities over
procurement of attacking progression. They are capable of forming a DNA complex termed as
Cas 1- Cas 2 protospacer when nominating a specific external DNA. Also, both the clusters
possess amid all the system of CRISPR Cas and introduce preservation that can be used in the
conditional genomic analysis. It has been determined that the new complex formed (Cas 1- Cas
2) behaves as an integrating agent novel spacer in CRISPR locus. Moreover, the latest spacers
are likely to aggregate into the end which is leading the duplicative spacer area (5’ area) although
latent spacers are suitable to be removed from the tail end of the duplicative spacer area (3’ end).
The occurrence of integration and deletion process can be simultaneous. Hence, the CRISPR
leading end displays high uncertainty while the CRISPR tail end shows high equality amid
various strains. Research determined that regularity among CRISPR and Cas protein is in or by
comparison extreme in similar type. This consideration signifies coevolution and interrelation of
mutual support among them. Also, there are critical similarities of various Cas protein amidst
different species, for instance, Cas 7 and Cas 9 all of which play an important part in protecting
external DNA. The research states that CRISPR 1 and CRISPR 4 are present in a few species
only while CRISPR is commonly found in most of the Streptococcus species.
Impact of the above system
11 | P a g e

Current researches regarding acquired immunization specified by CRISPR Cas system have
provided the fact that applying an efficient accession over S. thermophilus can protect it from
phages attack. The growth of S. thermophilus and reproduction is adversely affected by the S.
thermophilus phages and lead to a vast commercial breakdown in dairy production eventually. S.
thermophilus phages are divided into 2 parts namely pac and cos type in accordance with their
DNA cluster structure. The cos type part of S. thermophilus phages contains Sfi21, Sfi19, 7201,
and DT1 whereas the pac type phages contain Sfi11, 2972, and O1205. All of them are general
bacteriophages with the entire genome arranged and show huge similarities, which could help in
the improvement of the anti-phage capability of CRISPR Cas system.
The CRISPR Cas system frames a wall in bacteria that cannot be destroyed or broken, operating
through interrupting and cutting off external genes. When it is exposed to phage, a large group of
strains combines to form a progression from attacking phage as spacers. Moreover, the latest
spacer extension is discovered in CRISPR 1 and CRISPR 3 only. Although, there is no novel DR
progression acquired by CRISPR 2 and CRISPR 4. CRISPR 4 Cas exhibited biochemical actions
in vitro. After reduplication of the CRISPR locus consisting of extended DR progressions and
additional processing, the individual abandoned crRNA with a different architecture integrates
with Cas complex in order to design CRISPR effect nucleoprotein complex (crRNP). The main
function of crRNP is to work as endonuclease directed by crRNA. In the course of time, the
anchor part is invulnerable to the re-attacking phage by the way of sequence particular gap of
phage DNA (Gao, 2018).
The similarities among CRISPR spacer progressions and current progressions in GenBank
directory are examined and determined. First of all, most of the spacers are similar to S.
thermophilus bacteriophages genome. To the greatest extent, a single spacer can show
similarities with various phages and numerous spacers could oppose the equivalent phage.
Moreover, after studying the coevolution amid phages and hosts, it can be proposed or concluded
that, after disclosure to phage violation, new spacers are added to S. thermophilus CRISPR loci
as well as mutations or alterations may also take place in phage genome. Also, more recently
combined spacers arise from the classifying string of the managing genome (Luo, Leenay, and
Beisel, 2016).
12 | P a g e
provided the fact that applying an efficient accession over S. thermophilus can protect it from
phages attack. The growth of S. thermophilus and reproduction is adversely affected by the S.
thermophilus phages and lead to a vast commercial breakdown in dairy production eventually. S.
thermophilus phages are divided into 2 parts namely pac and cos type in accordance with their
DNA cluster structure. The cos type part of S. thermophilus phages contains Sfi21, Sfi19, 7201,
and DT1 whereas the pac type phages contain Sfi11, 2972, and O1205. All of them are general
bacteriophages with the entire genome arranged and show huge similarities, which could help in
the improvement of the anti-phage capability of CRISPR Cas system.
The CRISPR Cas system frames a wall in bacteria that cannot be destroyed or broken, operating
through interrupting and cutting off external genes. When it is exposed to phage, a large group of
strains combines to form a progression from attacking phage as spacers. Moreover, the latest
spacer extension is discovered in CRISPR 1 and CRISPR 3 only. Although, there is no novel DR
progression acquired by CRISPR 2 and CRISPR 4. CRISPR 4 Cas exhibited biochemical actions
in vitro. After reduplication of the CRISPR locus consisting of extended DR progressions and
additional processing, the individual abandoned crRNA with a different architecture integrates
with Cas complex in order to design CRISPR effect nucleoprotein complex (crRNP). The main
function of crRNP is to work as endonuclease directed by crRNA. In the course of time, the
anchor part is invulnerable to the re-attacking phage by the way of sequence particular gap of
phage DNA (Gao, 2018).
The similarities among CRISPR spacer progressions and current progressions in GenBank
directory are examined and determined. First of all, most of the spacers are similar to S.
thermophilus bacteriophages genome. To the greatest extent, a single spacer can show
similarities with various phages and numerous spacers could oppose the equivalent phage.
Moreover, after studying the coevolution amid phages and hosts, it can be proposed or concluded
that, after disclosure to phage violation, new spacers are added to S. thermophilus CRISPR loci
as well as mutations or alterations may also take place in phage genome. Also, more recently
combined spacers arise from the classifying string of the managing genome (Luo, Leenay, and
Beisel, 2016).
12 | P a g e
With the help of genome editing, that is done with the help of CRISPR Cas system, the diseases
that cause mutations or alteration in genes can also be cured. Some precise changes can be made
in the human body with the help of CRISPR Cas system.
The securing of new qualities that give a specifically preferred standpoint is a critical factor in
genome advancement. Extensive extents of archaeal and bacterial genomes comprise of genes
got from the trading of hereditary material among connected or disconnected species1, that is
termed as horizontal gene transfer (HGT). HGT happens by take-up of ecological DNA (change)
or by the fuse of different DNA carried on versatile hereditary components, for example,
bacteriophages (transduction) and plasmids (conjugation).
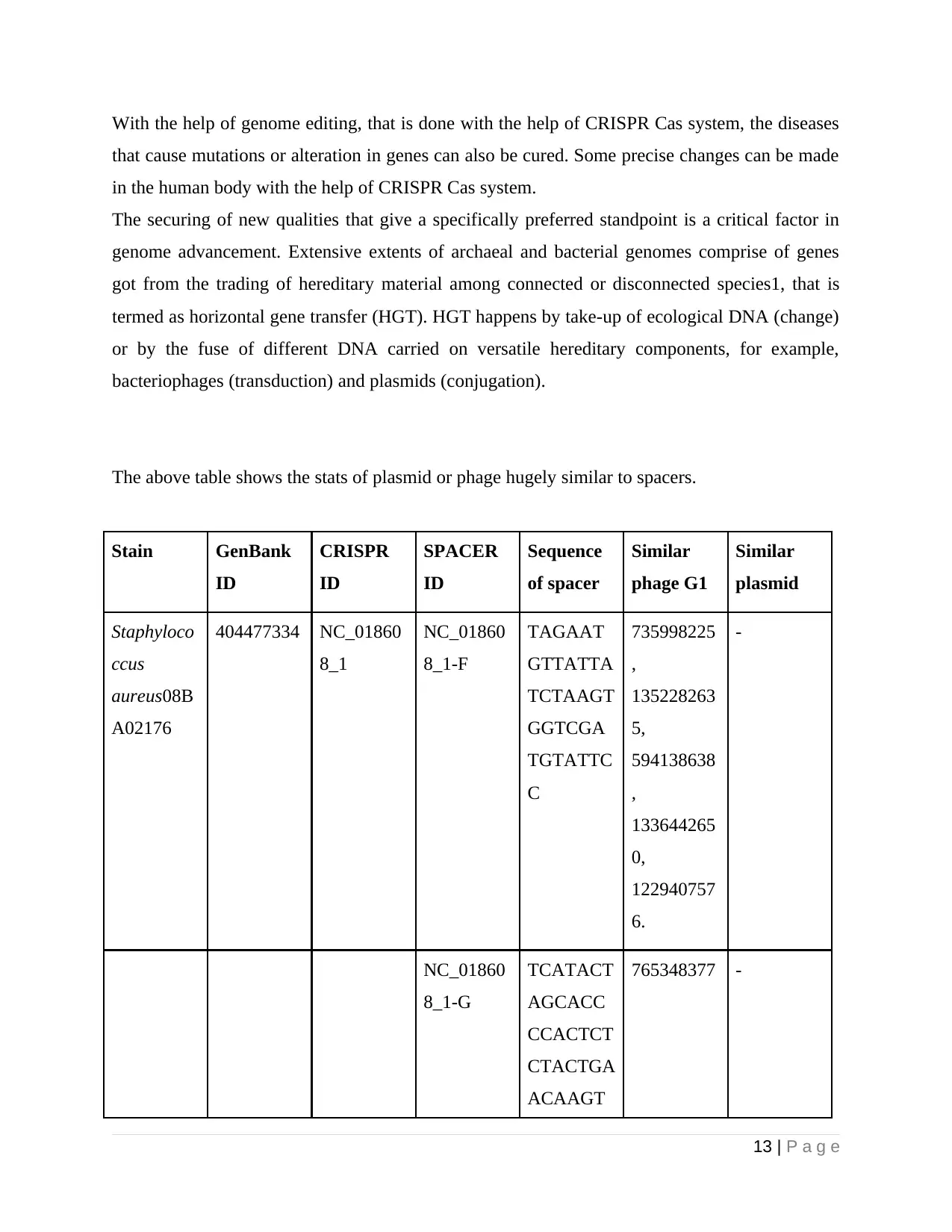
The above table shows the stats of plasmid or phage hugely similar to spacers.
Stain GenBank
ID
CRISPR
ID
SPACER
ID
Sequence
of spacer
Similar
phage G1
Similar
plasmid
Staphyloco
ccus
aureus08B
A02176
404477334 NC_01860
8_1
NC_01860
8_1-F
TAGAAT
GTTATTA
TCTAAGT
GGTCGA
TGTATTC
C
735998225
,
135228263
5,
594138638
,
133644265
0,
122940757
6.
-
NC_01860
8_1-G
TCATACT
AGCACC
CCACTCT
CTACTGA
ACAAGT
765348377 -
13 | P a g e
that cause mutations or alteration in genes can also be cured. Some precise changes can be made
in the human body with the help of CRISPR Cas system.
The securing of new qualities that give a specifically preferred standpoint is a critical factor in
genome advancement. Extensive extents of archaeal and bacterial genomes comprise of genes
got from the trading of hereditary material among connected or disconnected species1, that is
termed as horizontal gene transfer (HGT). HGT happens by take-up of ecological DNA (change)
or by the fuse of different DNA carried on versatile hereditary components, for example,
bacteriophages (transduction) and plasmids (conjugation).
The above table shows the stats of plasmid or phage hugely similar to spacers.
Stain GenBank
ID
CRISPR
ID
SPACER
ID
Sequence
of spacer
Similar
phage G1
Similar
plasmid
Staphyloco
ccus
aureus08B
A02176
404477334 NC_01860
8_1
NC_01860
8_1-F
TAGAAT
GTTATTA
TCTAAGT
GGTCGA
TGTATTC
C
735998225
,
135228263
5,
594138638
,
133644265
0,
122940757
6.
-
NC_01860
8_1-G
TCATACT
AGCACC
CCACTCT
CTACTGA
ACAAGT
765348377 -
13 | P a g e
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
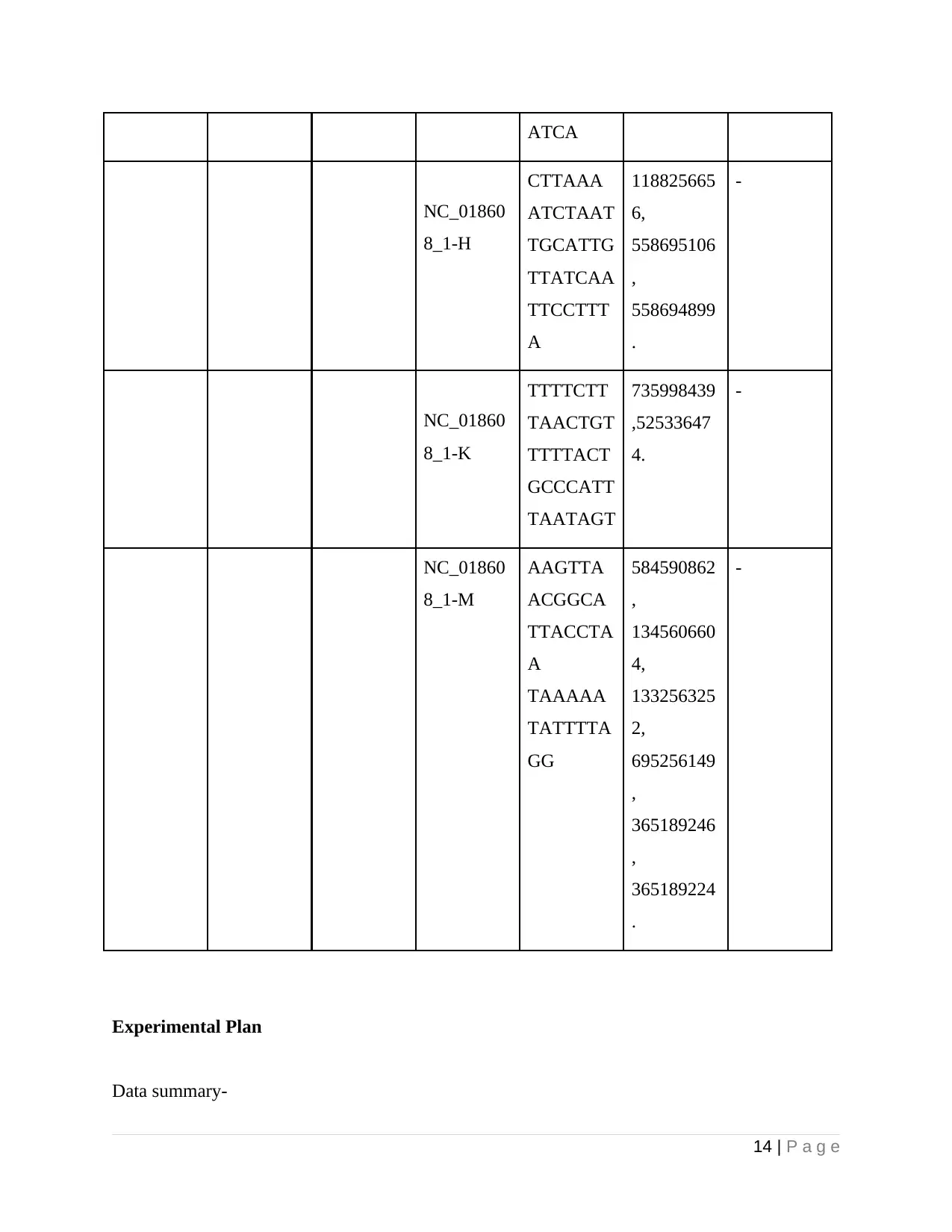
ATCA
NC_01860
8_1-H
CTTAAA
ATCTAAT
TGCATTG
TTATCAA
TTCCTTT
A
118825665
6,
558695106
,
558694899
.
-
NC_01860
8_1-K
TTTTCTT
TAACTGT
TTTTACT
GCCCATT
TAATAGT
735998439
,52533647
4.
-
NC_01860
8_1-M
AAGTTA
ACGGCA
TTACCTA
A
TAAAAA
TATTTTA
GG
584590862
,
134560660
4,
133256325
2,
695256149
,
365189246
,
365189224
.
-
Experimental Plan
Data summary-
14 | P a g e
NC_01860
8_1-H
CTTAAA
ATCTAAT
TGCATTG
TTATCAA
TTCCTTT
A
118825665
6,
558695106
,
558694899
.
-
NC_01860
8_1-K
TTTTCTT
TAACTGT
TTTTACT
GCCCATT
TAATAGT
735998439
,52533647
4.
-
NC_01860
8_1-M
AAGTTA
ACGGCA
TTACCTA
A
TAAAAA
TATTTTA
GG
584590862
,
134560660
4,
133256325
2,
695256149
,
365189246
,
365189224
.
-
Experimental Plan
Data summary-
14 | P a g e
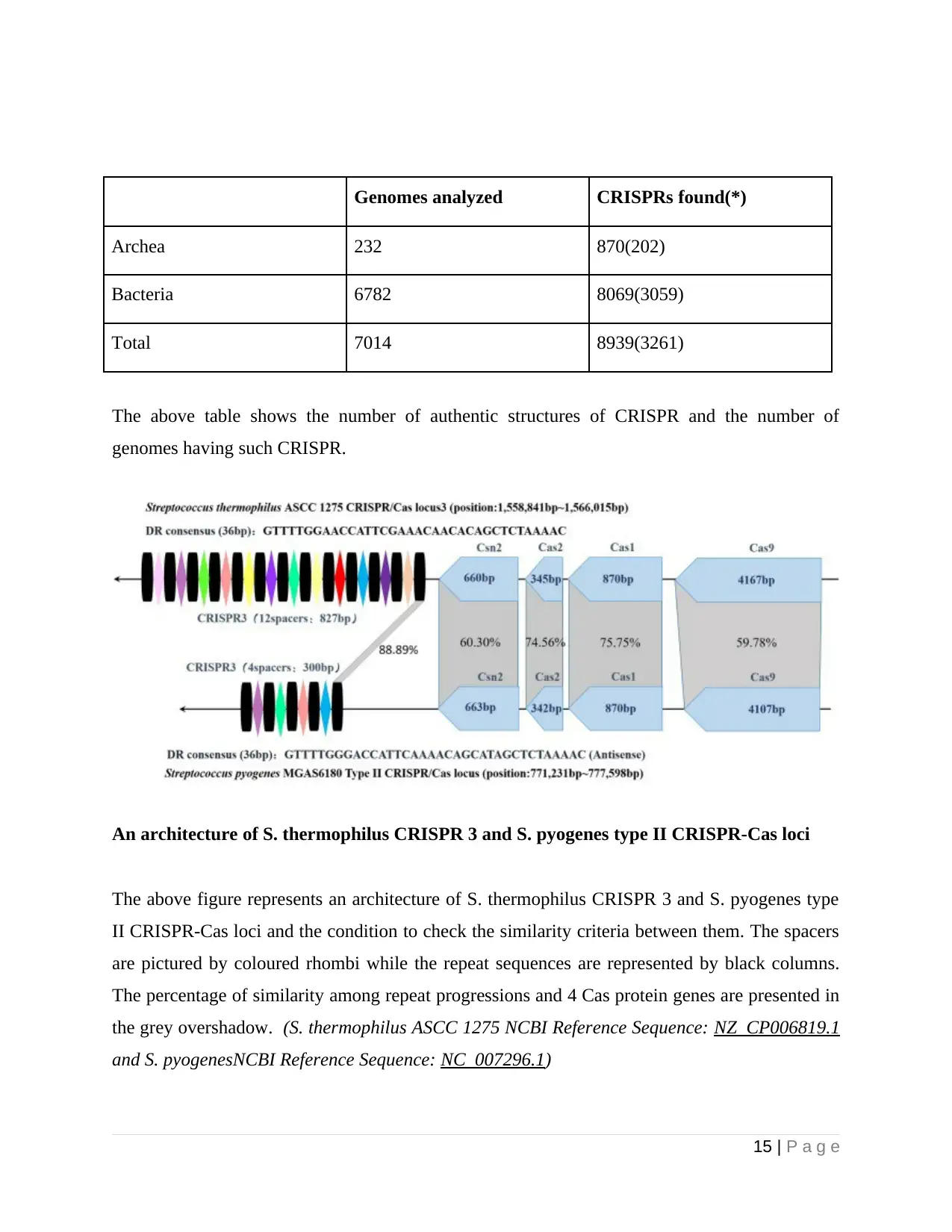
Genomes analyzed CRISPRs found(*)
Archea 232 870(202)
Bacteria 6782 8069(3059)
Total 7014 8939(3261)
The above table shows the number of authentic structures of CRISPR and the number of
genomes having such CRISPR.
An architecture of S. thermophilus CRISPR 3 and S. pyogenes type II CRISPR-Cas loci
The above figure represents an architecture of S. thermophilus CRISPR 3 and S. pyogenes type
II CRISPR-Cas loci and the condition to check the similarity criteria between them. The spacers
are pictured by coloured rhombi while the repeat sequences are represented by black columns.
The percentage of similarity among repeat progressions and 4 Cas protein genes are presented in
the grey overshadow. (S. thermophilus ASCC 1275 NCBI Reference Sequence: NZ_CP006819.1
and S. pyogenesNCBI Reference Sequence: NC_007296.1)
15 | P a g e
Archea 232 870(202)
Bacteria 6782 8069(3059)
Total 7014 8939(3261)
The above table shows the number of authentic structures of CRISPR and the number of
genomes having such CRISPR.
An architecture of S. thermophilus CRISPR 3 and S. pyogenes type II CRISPR-Cas loci
The above figure represents an architecture of S. thermophilus CRISPR 3 and S. pyogenes type
II CRISPR-Cas loci and the condition to check the similarity criteria between them. The spacers
are pictured by coloured rhombi while the repeat sequences are represented by black columns.
The percentage of similarity among repeat progressions and 4 Cas protein genes are presented in
the grey overshadow. (S. thermophilus ASCC 1275 NCBI Reference Sequence: NZ_CP006819.1
and S. pyogenesNCBI Reference Sequence: NC_007296.1)
15 | P a g e

From the above figure, it can be concluded that with the help of similarity comparison, the
duplicative progressions S. thermophilus CRISPR 3 and S. pyogenes type II CRISPR-Cas loci
use a common identity of 88.89% (32/36) with each other. Also, there are various spacers
combined within S. thermophilus CRISPR 3/Cas locus. Moreover, both of the systems of
CRISPR Cas exhibit an identical mechanism, having four Cas genes that include Cas 1, Cas 2,
Cas 9 and Csn 2 placed upstream and duplicative spacer progressions placed downstream the
loci. In addition to which, length of individual Cas gene is the same and the 4 parts of Cas
progressions exhibit relatively huge similarity. This is co-related with analogous actions. There
are further researches that prove that effective capability of S. thermophilus type II CRISPR-Cas
helps in the merging of external DNA. Of note, in the usual S. pyogenes CRISPR system, NGG
is used as its pAM loci even though S. themophilus Cas9 is capable of recognizing longer PAM.
the above aspect conveys St1Cas9 and St3Cas9 with greater addressing effectiveness and
progression particularity. Moreover, CRISPR Cas loci are major of 2 types S. pyogenes strains
genome that include type 1-C and type 2. Although, S. thermophilus CRISPR-Cas possesses
more ample varieties that includes type 3-A, type 1-E, and type 2-A. With respect to type 2-A
system, both the CRISPR 3 and CRISPR 1 systems are enriched with huge external DNA
opposition activity. Hence, S. thermophilus CRISPR-Cas system can perform better as well as it
can perform another characteristic miniature.
Portrait of a novel S. Thermophilus
CRISPR are hypervariable loci broadly conveyed in prokaryotes that give procured
insusceptibility against remote hereditary components. Here, we portray a novel Streptococcus
thermophilus locus which is termed as CRISPR 3 and tentatively exhibit its capacity to
coordinate novel spacers because of bacteriophage. Additionally, we break down CRISPR decent
variety and action crosswise over three particular CRISPR loci in a few S. thermophilus strains.
We demonstrate that both CRISPR rehashes and Cas genes are locus explicit and practically
coupled. An aggregate of 124 strains was examined, and 109 exceptional spacer plans were seen
over the three CRISPR loci. By and large, 3,626 spacers were investigated that included 2,829
for CRISPR 1 (782 one of a kind), a total of 173 for CRISPR 2 (16 special), and 624 for
CRISPR 3 (154 one of a kind). Arrangement investigation of the spacers uncovered similarity
16 | P a g e
duplicative progressions S. thermophilus CRISPR 3 and S. pyogenes type II CRISPR-Cas loci
use a common identity of 88.89% (32/36) with each other. Also, there are various spacers
combined within S. thermophilus CRISPR 3/Cas locus. Moreover, both of the systems of
CRISPR Cas exhibit an identical mechanism, having four Cas genes that include Cas 1, Cas 2,
Cas 9 and Csn 2 placed upstream and duplicative spacer progressions placed downstream the
loci. In addition to which, length of individual Cas gene is the same and the 4 parts of Cas
progressions exhibit relatively huge similarity. This is co-related with analogous actions. There
are further researches that prove that effective capability of S. thermophilus type II CRISPR-Cas
helps in the merging of external DNA. Of note, in the usual S. pyogenes CRISPR system, NGG
is used as its pAM loci even though S. themophilus Cas9 is capable of recognizing longer PAM.
the above aspect conveys St1Cas9 and St3Cas9 with greater addressing effectiveness and
progression particularity. Moreover, CRISPR Cas loci are major of 2 types S. pyogenes strains
genome that include type 1-C and type 2. Although, S. thermophilus CRISPR-Cas possesses
more ample varieties that includes type 3-A, type 1-E, and type 2-A. With respect to type 2-A
system, both the CRISPR 3 and CRISPR 1 systems are enriched with huge external DNA
opposition activity. Hence, S. thermophilus CRISPR-Cas system can perform better as well as it
can perform another characteristic miniature.
Portrait of a novel S. Thermophilus
CRISPR are hypervariable loci broadly conveyed in prokaryotes that give procured
insusceptibility against remote hereditary components. Here, we portray a novel Streptococcus
thermophilus locus which is termed as CRISPR 3 and tentatively exhibit its capacity to
coordinate novel spacers because of bacteriophage. Additionally, we break down CRISPR decent
variety and action crosswise over three particular CRISPR loci in a few S. thermophilus strains.
We demonstrate that both CRISPR rehashes and Cas genes are locus explicit and practically
coupled. An aggregate of 124 strains was examined, and 109 exceptional spacer plans were seen
over the three CRISPR loci. By and large, 3,626 spacers were investigated that included 2,829
for CRISPR 1 (782 one of a kind), a total of 173 for CRISPR 2 (16 special), and 624 for
CRISPR 3 (154 one of a kind). Arrangement investigation of the spacers uncovered similarity
16 | P a g e
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

and character to phage groupings (77%), plasmid successions (16%), and S. thermophilus
chromosomal arrangements (7%). Polymorphisms were watched for the CRISPR rehashes,
CRISPR spacers, Cas qualities, CRISPR theme, locus design, and explicit succession content.
Strangely, CRISPR loci developed both by means of captivated expansion of novel spacers after
introduction to remote hereditary components and through inner erasure of spacers. We guess
that the dimension of assorted variety is corresponded with relative CRISPR movement and
recommend that the action is most noteworthy for CRISPR1, trailed by CRISPR 3, while
CRISPR 2 might be degenerate. Universally, the dynamic idea of CRISPR loci may demonstrate
significant for composing and similar examinations of strains and microbial populaces. Likewise,
CRISPRs give basic bits of knowledge into the connections among prokaryotes and their
surroundings, strikingly the coevolution of viral and host genomes.
CRISPR and their related qualities are connected to an instrument of obtained obstruction against
bacteriophages. Microbes can incorporate short stretches of phage-determined arrangements
(spacers) inside CRISPR loci to wind up phage safe. In this examination, we further portrayed
the effectiveness of CRISPR 1 as a phage obstruction component in Streptococcus thermophilus.
To begin with, we demonstrate that CRISPR1 is unmistakable from recently known phage guard
frameworks and is compelling against the two fundamental gatherings of S. thermophilus
phages. Examinations of 30 bacteriophage-harsh freaks of S. thermophilus show that the
expansion of latest new spacer in CRISPR 1 is the most continuous result of a phage objection
and that the iterative expansion of spacers builds the general phage obstruction of the host. The
size of the new spacer added is considered to be between 29 to 31 nucleotides, with 30 being by
a wide margin the most incessant. A similar investigation of 39 recently procured spacers with
the total genomic successions of the wild-type phages 858, 2972, and DT1 exhibited that the
recently added spacer must be indistinguishable to an area (named proto-spacer) in the phage
genome to give a phage obstruction phenotype. In addition, we found a CRISPR 1-explicit
grouping (NNAGAAW) found downstream of the proto-spacer district that is essential for the
phage obstruction phenotype. At long last, we appear through the investigations of 20 freak
phages that destructive phages are quickly advancing through single nucleotide changes just as
erasures, in light of CRISPR 1 (Liu and Fan, 2014).
17 | P a g e
chromosomal arrangements (7%). Polymorphisms were watched for the CRISPR rehashes,
CRISPR spacers, Cas qualities, CRISPR theme, locus design, and explicit succession content.
Strangely, CRISPR loci developed both by means of captivated expansion of novel spacers after
introduction to remote hereditary components and through inner erasure of spacers. We guess
that the dimension of assorted variety is corresponded with relative CRISPR movement and
recommend that the action is most noteworthy for CRISPR1, trailed by CRISPR 3, while
CRISPR 2 might be degenerate. Universally, the dynamic idea of CRISPR loci may demonstrate
significant for composing and similar examinations of strains and microbial populaces. Likewise,
CRISPRs give basic bits of knowledge into the connections among prokaryotes and their
surroundings, strikingly the coevolution of viral and host genomes.
CRISPR and their related qualities are connected to an instrument of obtained obstruction against
bacteriophages. Microbes can incorporate short stretches of phage-determined arrangements
(spacers) inside CRISPR loci to wind up phage safe. In this examination, we further portrayed
the effectiveness of CRISPR 1 as a phage obstruction component in Streptococcus thermophilus.
To begin with, we demonstrate that CRISPR1 is unmistakable from recently known phage guard
frameworks and is compelling against the two fundamental gatherings of S. thermophilus
phages. Examinations of 30 bacteriophage-harsh freaks of S. thermophilus show that the
expansion of latest new spacer in CRISPR 1 is the most continuous result of a phage objection
and that the iterative expansion of spacers builds the general phage obstruction of the host. The
size of the new spacer added is considered to be between 29 to 31 nucleotides, with 30 being by
a wide margin the most incessant. A similar investigation of 39 recently procured spacers with
the total genomic successions of the wild-type phages 858, 2972, and DT1 exhibited that the
recently added spacer must be indistinguishable to an area (named proto-spacer) in the phage
genome to give a phage obstruction phenotype. In addition, we found a CRISPR 1-explicit
grouping (NNAGAAW) found downstream of the proto-spacer district that is essential for the
phage obstruction phenotype. At long last, we appear through the investigations of 20 freak
phages that destructive phages are quickly advancing through single nucleotide changes just as
erasures, in light of CRISPR 1 (Liu and Fan, 2014).
17 | P a g e

Moreover, with the help of different research, it is conveyed that by the application of genome
analysis on the 3 cos-type violent Streptococcus thermophilus phages that all of them are
equivalent to those unique in any another place in the world. Spontaneous bacteriophage-
insensitive S. thermophilus mutants (BIMs) were acquired after questioning the phage conscious
type of a wild strain Uy02 in association with the phage 128 and the analysis of their CRISPR
content was done. Examination of 23 BIMs clarified that all the phages access one unique spacer
in their respective CRISPR array. While the 5’ end space of the array is occupied by the other 14
BIMs and rest of the space left over with the array is occupied by the rest 9 BIMs. Examination
of the pioneer grouping in strains DGCC7710 and Uy02 demonstrated a nucleotide erasure at
position - 1 in Uy02, which might be in charge of the watched ectopic spacer procurement.
Investigation of the spacer successions upstream the recently gained ectopic spacer showed
nearness of a preserved adenine buildup at position - 2. This examination shows that normal
strains of S. thermophilus can likewise obtain spacers inside a CRISPR exhibit.
Summary
Streptococcus thermophilus CRISPR-Cas framework shows rich assorted varieties and fantastic
capacities of exogenous DNA mix. A lot of gainful qualities add to its splendid application
prospects. Attributable to the presence of RNA guided by Cas9 nucleases, S. thermophilus
CRISPR-Cas9 can be utilized as a productive genome alteration instrument with the assistance of
structured sgRNA. What's more, assorted variety and related development relationship give a one
of a kind point of view to arrangement and advancement investigates in Streptococcus and other
species which are co-related. Besides, phage contamination test in vitro together with similarity
investigation of spacers establish a framework for learns about the enemy of phage impact of
CRISPR-Cas framework. Future advancement of CRISPR-Cas will profit more from the
negative factors that influence its activities and development of its application go.
18 | P a g e
analysis on the 3 cos-type violent Streptococcus thermophilus phages that all of them are
equivalent to those unique in any another place in the world. Spontaneous bacteriophage-
insensitive S. thermophilus mutants (BIMs) were acquired after questioning the phage conscious
type of a wild strain Uy02 in association with the phage 128 and the analysis of their CRISPR
content was done. Examination of 23 BIMs clarified that all the phages access one unique spacer
in their respective CRISPR array. While the 5’ end space of the array is occupied by the other 14
BIMs and rest of the space left over with the array is occupied by the rest 9 BIMs. Examination
of the pioneer grouping in strains DGCC7710 and Uy02 demonstrated a nucleotide erasure at
position - 1 in Uy02, which might be in charge of the watched ectopic spacer procurement.
Investigation of the spacer successions upstream the recently gained ectopic spacer showed
nearness of a preserved adenine buildup at position - 2. This examination shows that normal
strains of S. thermophilus can likewise obtain spacers inside a CRISPR exhibit.
Summary
Streptococcus thermophilus CRISPR-Cas framework shows rich assorted varieties and fantastic
capacities of exogenous DNA mix. A lot of gainful qualities add to its splendid application
prospects. Attributable to the presence of RNA guided by Cas9 nucleases, S. thermophilus
CRISPR-Cas9 can be utilized as a productive genome alteration instrument with the assistance of
structured sgRNA. What's more, assorted variety and related development relationship give a one
of a kind point of view to arrangement and advancement investigates in Streptococcus and other
species which are co-related. Besides, phage contamination test in vitro together with similarity
investigation of spacers establish a framework for learns about the enemy of phage impact of
CRISPR-Cas framework. Future advancement of CRISPR-Cas will profit more from the
negative factors that influence its activities and development of its application go.
18 | P a g e

References-
Anders, C., Niewoehner, O., Duerst, A., and Jinek, M. (2014). Structural basis of PAM-
dependent target DNA recognition by the Cas9 endonuclease. Nature, 513(7519), pp. 569.
Barrangou, R. (2015). The diversity of CRISPR-Cas immune systems and molecular machines.
Genome biology, 16(1), 247.
Barrangou, R., and Doudna, J. A. (2016). Applications of CRISPR technologies in research and
beyond. Nature biotechnology, 34(9), pp. 933.
Bikard, D., and Marraffini, L. A. (2013). Control of gene expression by CRISPR-Cas systems.
F1000prime reports, 5.
Bikard, D., Jiang, W., Samai, P., Hochschild, A., Zhang, F., and Marraffini, L. A. (2013).
Programmable repression and activation of bacterial gene expression using an engineered
CRISPR-Cas system. Nucleic acids research, 41(15), pp. 7429-7437.
Blitz, I. L., Biesinger, J., Xie, X., and Cho, K. W. (2013). Biallelic genome modification in F0
Xenopus tropicalis embryos using the CRISPR/Cas system. genesis, 51(12), pp. 827-834.
Blosser, T. R., Loeff, L., Westra, E. R., Vlot, M., Künne, T., Sobota, M., ... and Joo, C. (2015).
Two distinct DNA binding modes guide the dual roles of a CRISPR-Cas protein complex.
Molecular Cell, 58(1), pp. 60-70.
Cho, J. S., Choi, K. R., Prabowo, C. P. S., Shin, J. H., Yang, D., Jang, J., and Lee, S. Y. (2017).
CRISPR/Cas9-coupled recombineering for metabolic engineering of Corynebacterium
glutamicum. Metabolic engineering, 42, pp. 157-167.
Chylinski, K., Makarova, K. S., Charpentier, E., and Koonin, E. V. (2014). Classification and
evolution of type II CRISPR-Cas systems. Nucleic acids research, 42(10), pp. 6091-6105.
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., ... and Zhang, F. (2013).
Multiplex genome engineering using CRISPR/Cas systems. Science, 339(6121), pp. 819-823.
Deveau, H., Barrangou, R., Garneau, J. E., Labonté, J., Fremaux, C., Boyaval, P., ... and
Moineau, S. (2009). Phage response to CRISPR-encoded resistance in Streptococcus
thermophilus. Journal of bacteriology, 190(4), pp.1390-1400.
Deveau, H., Garneau, J. E., and Moineau, S. (2010). CRISPR/Cas system and its role in phage-
bacteria interactions. Annual review of microbiology, 64, pp. 475-493.
19 | P a g e
Anders, C., Niewoehner, O., Duerst, A., and Jinek, M. (2014). Structural basis of PAM-
dependent target DNA recognition by the Cas9 endonuclease. Nature, 513(7519), pp. 569.
Barrangou, R. (2015). The diversity of CRISPR-Cas immune systems and molecular machines.
Genome biology, 16(1), 247.
Barrangou, R., and Doudna, J. A. (2016). Applications of CRISPR technologies in research and
beyond. Nature biotechnology, 34(9), pp. 933.
Bikard, D., and Marraffini, L. A. (2013). Control of gene expression by CRISPR-Cas systems.
F1000prime reports, 5.
Bikard, D., Jiang, W., Samai, P., Hochschild, A., Zhang, F., and Marraffini, L. A. (2013).
Programmable repression and activation of bacterial gene expression using an engineered
CRISPR-Cas system. Nucleic acids research, 41(15), pp. 7429-7437.
Blitz, I. L., Biesinger, J., Xie, X., and Cho, K. W. (2013). Biallelic genome modification in F0
Xenopus tropicalis embryos using the CRISPR/Cas system. genesis, 51(12), pp. 827-834.
Blosser, T. R., Loeff, L., Westra, E. R., Vlot, M., Künne, T., Sobota, M., ... and Joo, C. (2015).
Two distinct DNA binding modes guide the dual roles of a CRISPR-Cas protein complex.
Molecular Cell, 58(1), pp. 60-70.
Cho, J. S., Choi, K. R., Prabowo, C. P. S., Shin, J. H., Yang, D., Jang, J., and Lee, S. Y. (2017).
CRISPR/Cas9-coupled recombineering for metabolic engineering of Corynebacterium
glutamicum. Metabolic engineering, 42, pp. 157-167.
Chylinski, K., Makarova, K. S., Charpentier, E., and Koonin, E. V. (2014). Classification and
evolution of type II CRISPR-Cas systems. Nucleic acids research, 42(10), pp. 6091-6105.
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., ... and Zhang, F. (2013).
Multiplex genome engineering using CRISPR/Cas systems. Science, 339(6121), pp. 819-823.
Deveau, H., Barrangou, R., Garneau, J. E., Labonté, J., Fremaux, C., Boyaval, P., ... and
Moineau, S. (2009). Phage response to CRISPR-encoded resistance in Streptococcus
thermophilus. Journal of bacteriology, 190(4), pp.1390-1400.
Deveau, H., Garneau, J. E., and Moineau, S. (2010). CRISPR/Cas system and its role in phage-
bacteria interactions. Annual review of microbiology, 64, pp. 475-493.
19 | P a g e
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Gao, C., 2018. The future of CRISPR technologies in agriculture. Nat Rev Mol Cell Biol, 19(5),
pp.275-276.
Garneau, J. E., Dupuis, M. È., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., ... and
Moineau, S. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and
plasmid DNA. Nature, 468(7320), pp. 67.
Haeussler, M., and Concordet, J. P. (2016). Genome editing with CRISPR-Cas9: can it get any
better?. Journal of Genetics and Genomics, 43(5), pp. 239-250.
Horvath, P., Coûté-Monvoisin, A. C., Romero, D. A., Boyaval, P., Fremaux, C., and Barrangou,
R. (2009). Comparative analysis of CRISPR loci in lactic acid bacteria genomes. International
journal of food microbiology, 131(1), pp. 62-70.
Jiang, W., Bikard, D., Cox, D., Zhang, F., and Marraffini, L. A. (2013). RNA-guided editing of
bacterial genomes using CRISPR-Cas systems. Nature Biotechnology, 31(3),pp. 233.
Liu, L. and Fan, X.D., 2014. CRISPR–Cas system: a powerful tool for genome
engineering. Plant molecular biology, 85(3), pp.209-218.
Lü, J., Chen, G., Cheng, D., and Celikovsky, S. (2002). Bridge the gap between the Lorenz
system and the Chen system. International Journal of Bifurcation and Chaos, 12(12), pp. 2917-
2926.
Lu, Y., and Zhu, J. K. (2017). Precise editing of a target base in the rice genome using a
modified CRISPR/Cas9 system. Molecular plant, 10(3), pp. 523-525.
Luo, M.L., Leenay, R.T. and Beisel, C.L., 2016. Current and future prospects for CRISPR‐based
tools in bacteria. Biotechnology and bioengineering, 113(5), pp.930-943.
Ma, X., Zhu, Q., Chen, Y., and Liu, Y. G. (2016). CRISPR/Cas9 platforms for genome editing in
plants: developments and applications. Molecular plant, 9(7), pp. 961-974.
Magadán, A. H., Dupuis, M. È., Villion, M., and Moineau, S. (2012). Cleavage of phage DNA
by the Streptococcus thermophilus CRISPR3-Cas system. PloS one, 7(7),p. e40913.
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J., Charpentier, E., Horvath, P., ... and
Van Der Oost, J. (2011). Evolution and classification of the CRISPR–Cas systems. Nature
Reviews Microbiology, 9(6), pp. 467.
Mandal, P. K., Ferreira, L. M., Collins, R., Meissner, T. B., Boutwell, C. L., Friesen, M., ... and
Musunuru, K. (2014). Efficient ablation of genes in human hematopoietic stem and effector cells
using CRISPR/Cas9. Cell stem cell, 15(5),pp. 643-652.
20 | P a g e
pp.275-276.
Garneau, J. E., Dupuis, M. È., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., ... and
Moineau, S. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and
plasmid DNA. Nature, 468(7320), pp. 67.
Haeussler, M., and Concordet, J. P. (2016). Genome editing with CRISPR-Cas9: can it get any
better?. Journal of Genetics and Genomics, 43(5), pp. 239-250.
Horvath, P., Coûté-Monvoisin, A. C., Romero, D. A., Boyaval, P., Fremaux, C., and Barrangou,
R. (2009). Comparative analysis of CRISPR loci in lactic acid bacteria genomes. International
journal of food microbiology, 131(1), pp. 62-70.
Jiang, W., Bikard, D., Cox, D., Zhang, F., and Marraffini, L. A. (2013). RNA-guided editing of
bacterial genomes using CRISPR-Cas systems. Nature Biotechnology, 31(3),pp. 233.
Liu, L. and Fan, X.D., 2014. CRISPR–Cas system: a powerful tool for genome
engineering. Plant molecular biology, 85(3), pp.209-218.
Lü, J., Chen, G., Cheng, D., and Celikovsky, S. (2002). Bridge the gap between the Lorenz
system and the Chen system. International Journal of Bifurcation and Chaos, 12(12), pp. 2917-
2926.
Lu, Y., and Zhu, J. K. (2017). Precise editing of a target base in the rice genome using a
modified CRISPR/Cas9 system. Molecular plant, 10(3), pp. 523-525.
Luo, M.L., Leenay, R.T. and Beisel, C.L., 2016. Current and future prospects for CRISPR‐based
tools in bacteria. Biotechnology and bioengineering, 113(5), pp.930-943.
Ma, X., Zhu, Q., Chen, Y., and Liu, Y. G. (2016). CRISPR/Cas9 platforms for genome editing in
plants: developments and applications. Molecular plant, 9(7), pp. 961-974.
Magadán, A. H., Dupuis, M. È., Villion, M., and Moineau, S. (2012). Cleavage of phage DNA
by the Streptococcus thermophilus CRISPR3-Cas system. PloS one, 7(7),p. e40913.
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J., Charpentier, E., Horvath, P., ... and
Van Der Oost, J. (2011). Evolution and classification of the CRISPR–Cas systems. Nature
Reviews Microbiology, 9(6), pp. 467.
Mandal, P. K., Ferreira, L. M., Collins, R., Meissner, T. B., Boutwell, C. L., Friesen, M., ... and
Musunuru, K. (2014). Efficient ablation of genes in human hematopoietic stem and effector cells
using CRISPR/Cas9. Cell stem cell, 15(5),pp. 643-652.
20 | P a g e

Puchta, H. (2016). Using CRISPR/Cas in three dimensions: towards synthetic plant genomes,
transcriptomes and epigenomes. The plant journal, 87(1), pp. 5-15.
Ran, F. A., Hsu, P. D., Lin, C. Y., Gootenberg, J. S., Konermann, S., Trevino, A. E., ... and
Zhang, F. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing
specificity. Cell, 154(6), pp. 1380-1389.
Sanozky-Dawes, R., Selle, K., O'Flaherty, S., Klaenhammer, T., and Barrangou, R. (2015).
Occurrence and activity of a type II CRISPR-Cas system in Lactobacillus gasseri. Microbiology,
161(9), pp. 1752-1761.
Selle, K., and Barrangou, R. (2015). CRISPR‐Based Technologies and the Future of Food
Science. Journal of food science, 80(11), pp. R2367-R2372.
Selle, K., and Barrangou, R. (2015). Harnessing CRISPR–Cas systems for bacterial genome
editing. Trends in microbiology, 23(4), pp. 225-232.
Selle, K., Klaenhammer, T. R., and Barrangou, R. (2015). CRISPR-based screening of genomic
island excision events in bacteria. Proceedings of the National Academy of Sciences, 112(26),pp.
8076-8081.
Sharan, S. K., Thomason, L. C., Kuznetsov, S. G., and Court, D. L. (2009). Recombineering: a
homologous recombination-based method of genetic engineering. Nature protocols, 4(2), pp.
206.
Tian, S., Jiang, L., Gao, Q., Zhang, J., Zong, M., Zhang, H., ... and Xu, Y. (2017). Efficient
CRISPR/Cas9-based gene knockout in watermelon. Plant cell reports, 36(3), pp. 399-406.
Wang, Y., Zhang, Z. T., Seo, S. O., Choi, K., Lu, T., Jin, Y. S., and Blaschek, H. P. (2015).
Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system.
Journal of Biotechnology, 200, pp. 1-5.
Wei, Y., Terns, R. M., and Terns, M. P. (2015). Cas9 function and host genome sampling in
Type II-A CRISPR–Cas adaptation. Genes and development, 29(4), pp. 356-361.
Zhu, G. H., Xu, J., Cui, Z., Dong, X. T., Ye, Z. F., Niu, D. J., ... and Dong, S. L. (2016).
Functional characterization of SlitPBP3 in Spodoptera litura by CRISPR/Cas9 mediated genome
editing. Insect biochemistry and molecular biology, pp. 75, 1-9.
21 | P a g e
transcriptomes and epigenomes. The plant journal, 87(1), pp. 5-15.
Ran, F. A., Hsu, P. D., Lin, C. Y., Gootenberg, J. S., Konermann, S., Trevino, A. E., ... and
Zhang, F. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing
specificity. Cell, 154(6), pp. 1380-1389.
Sanozky-Dawes, R., Selle, K., O'Flaherty, S., Klaenhammer, T., and Barrangou, R. (2015).
Occurrence and activity of a type II CRISPR-Cas system in Lactobacillus gasseri. Microbiology,
161(9), pp. 1752-1761.
Selle, K., and Barrangou, R. (2015). CRISPR‐Based Technologies and the Future of Food
Science. Journal of food science, 80(11), pp. R2367-R2372.
Selle, K., and Barrangou, R. (2015). Harnessing CRISPR–Cas systems for bacterial genome
editing. Trends in microbiology, 23(4), pp. 225-232.
Selle, K., Klaenhammer, T. R., and Barrangou, R. (2015). CRISPR-based screening of genomic
island excision events in bacteria. Proceedings of the National Academy of Sciences, 112(26),pp.
8076-8081.
Sharan, S. K., Thomason, L. C., Kuznetsov, S. G., and Court, D. L. (2009). Recombineering: a
homologous recombination-based method of genetic engineering. Nature protocols, 4(2), pp.
206.
Tian, S., Jiang, L., Gao, Q., Zhang, J., Zong, M., Zhang, H., ... and Xu, Y. (2017). Efficient
CRISPR/Cas9-based gene knockout in watermelon. Plant cell reports, 36(3), pp. 399-406.
Wang, Y., Zhang, Z. T., Seo, S. O., Choi, K., Lu, T., Jin, Y. S., and Blaschek, H. P. (2015).
Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system.
Journal of Biotechnology, 200, pp. 1-5.
Wei, Y., Terns, R. M., and Terns, M. P. (2015). Cas9 function and host genome sampling in
Type II-A CRISPR–Cas adaptation. Genes and development, 29(4), pp. 356-361.
Zhu, G. H., Xu, J., Cui, Z., Dong, X. T., Ye, Z. F., Niu, D. J., ... and Dong, S. L. (2016).
Functional characterization of SlitPBP3 in Spodoptera litura by CRISPR/Cas9 mediated genome
editing. Insect biochemistry and molecular biology, pp. 75, 1-9.
21 | P a g e
1 out of 21
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
© 2024 | Zucol Services PVT LTD | All rights reserved.

