Protein Chemistry Report: Analysis of Fu et al. 2020 Article
VerifiedAdded on 2022/01/15
|5
|2715
|101
Report
AI Summary
This report provides a comprehensive review of the article by Fu et al. (2020) concerning the influence of the exogenous Cry1Ab/c protein on endogenous proteins in transgenic rice. The study investigates protein-protein interactions, employing techniques such as yeast two-hybrid assays, subcellular co-localization, bimolecular fluorescence complementation (BiFC), and co-immunoprecipitation (co-IP). The report critically assesses the methodologies used, highlighting potential biases and suggesting improvements, such as the use of protoplast two-hybrid assays and label transfer screening to enhance the accuracy and efficiency of interaction analysis. The report also discusses the importance of comprehensive data presentation, including complete SDS-PAGE gels with controls, to ensure unambiguous interpretation of results. The article's findings suggest that the exogenous protein interacts with endogenous proteins involved in stress resistance and photosynthesis, leading to phenotypic changes, and the report evaluates the strengths and weaknesses of the experimental approaches used to validate these interactions.

ARTICLE ASSIGNMENT:
FU ET AL. 2020
PROTEIN CHEMISTRY
Prof. dr. Els Van Damme
Dr. Tibo De Coninck
Joppe Gebruers
Master cell and gene biotechnology
Year: 2021-2022
FU ET AL. 2020
PROTEIN CHEMISTRY
Prof. dr. Els Van Damme
Dr. Tibo De Coninck
Joppe Gebruers
Master cell and gene biotechnology
Year: 2021-2022
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

1. INTRODUCTION
This report reviews the article on the influence (plant growth and development) of exogenous Cry1Ab/c
protein on endogenous proteins from Fu, et al. (2020), is discussed.[1] Genes, which are not inherently
present in the genome (exogenous), are added to the genetic code, hence forming transgenic crops.
Making these transgenic organisms can have several goals e.g., higher yield to cost ratio, making it
economically beneficial to use these ‘enhanced’ crops. Therefore, the interest in these transgenic crops
is rising over the years. However, these exogenous proteins can interact with the already present
endogenous proteins, inducing unintended responses. A previous study, by the same group, showed that
the insect-resistant transgenic rice (Huahui-1) differs vastly (in plant height, biomass, etc.) from the
parental rice.[2] In the reviewed article, these protein-protein interactions are examined with the use of
Cry1Ab/c (protein from the insect-resistant rice Huahui-1) and the endogenous proteins of Minghui-63
(the parental rice). Fu, et al. (2020) suspect that the expressed CryAb/c in Huahui-1 interacts with
endogenous proteins, resulting in the previously observed phenotypical changes.
To analyze these interactions, multiple techniques are used. Firstly, a yeast two-hybrid assay is used to
get an idea of which endogenous proteins will interact with the Cry1Ab/c. 60 Proteins are identified as
Cry1Ab/c-interacting endogenous proteins. The researchers only focus on proteins involved in stress
resistance and photosynthesis assuming that these two factors lay on the basis of the phenotypical
difference. This premise provided a library reduction resulting in 14 interesting (and interacting)
endogenous proteins. For further identification and analysis of the interactions taking place, three
validation methods were used: subcellular co-localization, bimolecular fluorescence complementation
(BIFC) and co-immunoprecipitation (co-IP). The results of these three techniques were used to make
conclusions about the influence of the exogenous protein.
This report focuses on the protein chemistry side of the article, discussing the used techniques and
whether improvements are possible. The gene technological side of the research (e.g., the use of a
certain vector or a certain yeast species) is not discussed further.
2. USED TECHNIQUES
The first technique used in the article is the yeast two-hybrid assay (Y2H) to identify which endogenous
proteins will interact with the Cry1Ab/c protein. This system is based on a split transcription factor that is
reconstructed when two proteins interact with one another. The bait protein (in this case Cry1Ab/c) is
linked to a DNA binding domain, making it able to bind DNA specifically on a promotor sequence. The
prey on the other hand is linked with an activation domain, making it able to activate transcription of a
certain reporter gene. This can only occur if the activation domain is close enough to the gene.
Therefore, the full transcription factor (DNA binding domain and activation domain) needs to be present
to activate transcription. Only if the bait protein interacts with the prey protein, this will take place.
However, two potential pitfalls of this assay need to be verified. False positives can occur due to self-
activation of the bait protein, or no activation can occur due to cytotoxicity of the bait protein. These two
concerns were analyzed critically in the article. As a matter of fact, the former proves to be present. To
solve this issue, the researchers cleaved the Cry1Ab/c into three segments dependent on their separate
activity. These three domain-BD constructs were again tested on self-activation and cytotoxicity,
resulting in a doubly negative result. Thus, these constructs can be used to identify endogenous proteins,
adding them as bait for the Y2H screening. The assay resulted in 60 positive clones (meaning interacting
endogenous protein), but after selection for photosynthesis and stress resistance proteins, the library
This report reviews the article on the influence (plant growth and development) of exogenous Cry1Ab/c
protein on endogenous proteins from Fu, et al. (2020), is discussed.[1] Genes, which are not inherently
present in the genome (exogenous), are added to the genetic code, hence forming transgenic crops.
Making these transgenic organisms can have several goals e.g., higher yield to cost ratio, making it
economically beneficial to use these ‘enhanced’ crops. Therefore, the interest in these transgenic crops
is rising over the years. However, these exogenous proteins can interact with the already present
endogenous proteins, inducing unintended responses. A previous study, by the same group, showed that
the insect-resistant transgenic rice (Huahui-1) differs vastly (in plant height, biomass, etc.) from the
parental rice.[2] In the reviewed article, these protein-protein interactions are examined with the use of
Cry1Ab/c (protein from the insect-resistant rice Huahui-1) and the endogenous proteins of Minghui-63
(the parental rice). Fu, et al. (2020) suspect that the expressed CryAb/c in Huahui-1 interacts with
endogenous proteins, resulting in the previously observed phenotypical changes.
To analyze these interactions, multiple techniques are used. Firstly, a yeast two-hybrid assay is used to
get an idea of which endogenous proteins will interact with the Cry1Ab/c. 60 Proteins are identified as
Cry1Ab/c-interacting endogenous proteins. The researchers only focus on proteins involved in stress
resistance and photosynthesis assuming that these two factors lay on the basis of the phenotypical
difference. This premise provided a library reduction resulting in 14 interesting (and interacting)
endogenous proteins. For further identification and analysis of the interactions taking place, three
validation methods were used: subcellular co-localization, bimolecular fluorescence complementation
(BIFC) and co-immunoprecipitation (co-IP). The results of these three techniques were used to make
conclusions about the influence of the exogenous protein.
This report focuses on the protein chemistry side of the article, discussing the used techniques and
whether improvements are possible. The gene technological side of the research (e.g., the use of a
certain vector or a certain yeast species) is not discussed further.
2. USED TECHNIQUES
The first technique used in the article is the yeast two-hybrid assay (Y2H) to identify which endogenous
proteins will interact with the Cry1Ab/c protein. This system is based on a split transcription factor that is
reconstructed when two proteins interact with one another. The bait protein (in this case Cry1Ab/c) is
linked to a DNA binding domain, making it able to bind DNA specifically on a promotor sequence. The
prey on the other hand is linked with an activation domain, making it able to activate transcription of a
certain reporter gene. This can only occur if the activation domain is close enough to the gene.
Therefore, the full transcription factor (DNA binding domain and activation domain) needs to be present
to activate transcription. Only if the bait protein interacts with the prey protein, this will take place.
However, two potential pitfalls of this assay need to be verified. False positives can occur due to self-
activation of the bait protein, or no activation can occur due to cytotoxicity of the bait protein. These two
concerns were analyzed critically in the article. As a matter of fact, the former proves to be present. To
solve this issue, the researchers cleaved the Cry1Ab/c into three segments dependent on their separate
activity. These three domain-BD constructs were again tested on self-activation and cytotoxicity,
resulting in a doubly negative result. Thus, these constructs can be used to identify endogenous proteins,
adding them as bait for the Y2H screening. The assay resulted in 60 positive clones (meaning interacting
endogenous protein), but after selection for photosynthesis and stress resistance proteins, the library

was shrunk down to 14 proteins. These will further be analyzed by respectively subcellular co-
localization, BIFC and co-IP.
Subcellular co-localization is used to determine the location of a protein within a cell with the use of
fluorescent markers. In this case, the Cry1Ab/c (full-length) is linked to mCherry fluorescent protein,
whereas the 14 endogenous proteins were linked to green fluorescent protein (GFP). The assay showed
that the exogenous protein mainly occurs in the nucleus and cytoplasm. All the endogenous proteins co-
localized with the Cry1Ab/c-mCherry, with locations differing between the proteins. The majority of
photosynthetic proteins were found co-localizing in the chloroplast, where the stress resistance proteins
mostly co-localized in the cytoplasm and nucleus. To check for co-localization two separate fluorescent
images are merged. Here the Cry1Ab/c protein is found in the red sites (linked to mCherry) and the
exogenous proteins are found in the green sites (linked to GFP).[3] A yellow to orange color gradient is
observed if the two proteins are in close proximity to one another.
Bimolecular Fluorescence Complementation (abbreviated BiFC) is a technique used to check in-vivo
protein-protein interactions. To accomplish this, YFP (Yellow Fluorescent Protein) is split into two
fragments: an N-terminal and a C-terminal half. The former is linked to the full-length Cry1Ab/c, whereas
the latter is fused to the 14 endogenous rice proteins. Both constructs are inserted in an expression
vector and expressed in tobacco mesophyll cells. If two proteins would interact, the two YFP-fragments
can fuse, hence a yellow fluorescent signal from a restored YFP. Protein-protein interactions (intensity
and location) are determined by the intensity of YFP fluorescence. An important downside of using BiFC
is the fact that it is prone to false-positive results due to spontaneous fluorescence. Therefore, a second
technique (co-IP) is used to confirm the protein interactions. Furthermore, the addition of an extra
fragment to the interacting proteins can lead to steric hindrance and thus interfere with the very specific
interaction.[4] Fu, et al. (2020) does not elaborate on this constraint, leading to potential false-negative
results (no protein interaction due to steric hindrance).
Co-immunoprecipitation (abbreviated co-IP) is the fourth and last technique used to confirm the
interactions between proteins. In a normal immunoprecipitation (IP), an antibody will bind specifically to
the protein of interest in a mixture of proteins. By binding the antibody to a second protein (protein G or
A, making the complex insoluble), the antibody-protein construct precipitates after centrifugation. Fu , et
al. (2020) uses a co-IP so that, together with the protein of interest, the interacting proteins precipitate
as well. In this case, anti-GFP antibodies are used to detect the target endogenous protein whereas anti-
mCherry antibodies were used to detect Cry1Ab/c. The results were visualized with the use of SDS-PAGE.
For this, a protein extraction is needed. Fu, et al. (2020) does not mention the contents of the used
extraction buffer. Only the used detergent is indicated, being Triton (20%). Also here, no further
information is given about which Triton detergent is used. Both BiFC and co-IP grant information about
the interaction between the 14 endogenous proteins and Cry1Ab/c, therefore both results can be
compared.
3. DISCUSSION
Fu, et al. (2020) uses four different techniques to assess the influence of exogenous protein on the
growth and development of a plant. In this case, Cry1Ab/c exogenous protein is expressed in Huahui-1
transgenic rice. They assume that proteins involved in stress resistance or photosynthesis are responsible
for the phenotypical change caused by Cry1Ab/c. They conclude that the exogenous protein interacts
with 60 endogenous proteins of which 14 are identified as being involved in stress resistance and
localization, BIFC and co-IP.
Subcellular co-localization is used to determine the location of a protein within a cell with the use of
fluorescent markers. In this case, the Cry1Ab/c (full-length) is linked to mCherry fluorescent protein,
whereas the 14 endogenous proteins were linked to green fluorescent protein (GFP). The assay showed
that the exogenous protein mainly occurs in the nucleus and cytoplasm. All the endogenous proteins co-
localized with the Cry1Ab/c-mCherry, with locations differing between the proteins. The majority of
photosynthetic proteins were found co-localizing in the chloroplast, where the stress resistance proteins
mostly co-localized in the cytoplasm and nucleus. To check for co-localization two separate fluorescent
images are merged. Here the Cry1Ab/c protein is found in the red sites (linked to mCherry) and the
exogenous proteins are found in the green sites (linked to GFP).[3] A yellow to orange color gradient is
observed if the two proteins are in close proximity to one another.
Bimolecular Fluorescence Complementation (abbreviated BiFC) is a technique used to check in-vivo
protein-protein interactions. To accomplish this, YFP (Yellow Fluorescent Protein) is split into two
fragments: an N-terminal and a C-terminal half. The former is linked to the full-length Cry1Ab/c, whereas
the latter is fused to the 14 endogenous rice proteins. Both constructs are inserted in an expression
vector and expressed in tobacco mesophyll cells. If two proteins would interact, the two YFP-fragments
can fuse, hence a yellow fluorescent signal from a restored YFP. Protein-protein interactions (intensity
and location) are determined by the intensity of YFP fluorescence. An important downside of using BiFC
is the fact that it is prone to false-positive results due to spontaneous fluorescence. Therefore, a second
technique (co-IP) is used to confirm the protein interactions. Furthermore, the addition of an extra
fragment to the interacting proteins can lead to steric hindrance and thus interfere with the very specific
interaction.[4] Fu, et al. (2020) does not elaborate on this constraint, leading to potential false-negative
results (no protein interaction due to steric hindrance).
Co-immunoprecipitation (abbreviated co-IP) is the fourth and last technique used to confirm the
interactions between proteins. In a normal immunoprecipitation (IP), an antibody will bind specifically to
the protein of interest in a mixture of proteins. By binding the antibody to a second protein (protein G or
A, making the complex insoluble), the antibody-protein construct precipitates after centrifugation. Fu , et
al. (2020) uses a co-IP so that, together with the protein of interest, the interacting proteins precipitate
as well. In this case, anti-GFP antibodies are used to detect the target endogenous protein whereas anti-
mCherry antibodies were used to detect Cry1Ab/c. The results were visualized with the use of SDS-PAGE.
For this, a protein extraction is needed. Fu, et al. (2020) does not mention the contents of the used
extraction buffer. Only the used detergent is indicated, being Triton (20%). Also here, no further
information is given about which Triton detergent is used. Both BiFC and co-IP grant information about
the interaction between the 14 endogenous proteins and Cry1Ab/c, therefore both results can be
compared.
3. DISCUSSION
Fu, et al. (2020) uses four different techniques to assess the influence of exogenous protein on the
growth and development of a plant. In this case, Cry1Ab/c exogenous protein is expressed in Huahui-1
transgenic rice. They assume that proteins involved in stress resistance or photosynthesis are responsible
for the phenotypical change caused by Cry1Ab/c. They conclude that the exogenous protein interacts
with 60 endogenous proteins of which 14 are identified as being involved in stress resistance and
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

photosynthesis. A combination of these 14 interactions can result in a changing phenotype.
The yeast two-hybrid screening is performed considering two major biases: cytotoxicity and self-
activation. These can lead to false negative and false positive results respectively. Both biases were
resolved by cleaving the Cry1Ab/c protein into three segments. Although this solves the problem with
self-activation, the original function will also be drastically impacted or completely lost. For this reason,
the full-length protein is used to determine further interactions with the other techniques. A third
drawback of using the yeast two-hybrid system is inherent to the technique. A yeast cell differs
considerably from a plant cell in terms of protein content and modification.[5] In conclusion is the Y2H
assay a very powerful method to suspect interactions but has the drawback of having to deal with a large
number of false positive and false negative results.[6] Therefore, Fu, et al. (2020) does not rely on this
technique to draw conclusions and exerted two other analyses (co-IP and BiFC). As an alternative for this
technique, a protoplast two-hybrid (P2H) assay can be used. A protoplast that resembles the plant cell of
interest is then used as the host for the two-hybrid screening. Ehlert, et al. proved this technique to be a
good alternative and tool for in vivo protein-protein interaction analysis.[7]
The results of the Y2H screening were first validated by a subcellular co-localization test. Based on the
differences between the original location of the endogenous proteins and the locations of interaction,
speculations about the influence of Cry1Ab/c were made.
To check if the proteins do indeed interact with the full-length Cry1Ab/c, a BiFC analysis is performed.
The interactions between proteins are very specific and prone to small differences in structure. As
already mentioned, the addition of an extra fragment to the interacting protein can lead to steric
hindrance and thus lead to failing interactions. To overcome this, multiple constructs must be made to
cover ideally every possible interaction manner. Fu, et al. (2020) chooses to only cover one combination
of constructs, fusing the N-terminal half to Cry1Ab/c and the C-terminal half to the interacting proteins.
To get a higher quality result, more constructs must be made.[4] To avoid this higher workload, a label
transfer screening can be used instead[8]. This uses a label transfer reagent (LTR) fused to one of the
interacting proteins. As it interacts with another protein, the LTR folds back leading to a second
interaction. As a result, the second protein now contains the photoreactive site proving the interaction
that just took place. The big advantage of using this over BiFC is that only one protein must be linked to
the LTR so that no combinations of constructs need to be made. Also, the LTR is structurally much
smaller than half of YFP, making for a lower chance of steric hindrance.
Co-IP is used as a second technique to confirm the interactions between proteins found by Y2H. As
already mentioned, is the BiFC analysis prone to spontaneous fluorescence leading to false positives,
hence the need for a second technique. The results for co-IP are largely in accordance with those from
BiFC except for one endogenous protein (CP43). According to the BiFC assay, it would interact with
Cry1Ab/c as opposed to the result of co-IP, suggesting a false positive BiFC result. The co-IP is analyzed
via SDS-PAGE, which results in Figure 1. A first critical note must be made about the lack of information
about the used extraction method. It is not indicated which buffer and protocol to use. This makes it
more difficult to get comparable results for other research. The SDS-PAGE gel itself is cut up into small
images covering only the regions where the expected bands are visible. The writers indicate the targeted
bands with a red asterisk instead of adding images of the full SDS-PAGE gels, including the used ladder
and controls. This would make for a more standardized result and unambiguous interpretation of the
data, as now there is no context to the given pictures.
To conclude, Fu, et al. (2020) uses 4 techniques to inspect the influence of Cry1Ab/c on endogenous
proteins. They use yeast two-hybrid screening to get a library of all interacting proteins and picked the
The yeast two-hybrid screening is performed considering two major biases: cytotoxicity and self-
activation. These can lead to false negative and false positive results respectively. Both biases were
resolved by cleaving the Cry1Ab/c protein into three segments. Although this solves the problem with
self-activation, the original function will also be drastically impacted or completely lost. For this reason,
the full-length protein is used to determine further interactions with the other techniques. A third
drawback of using the yeast two-hybrid system is inherent to the technique. A yeast cell differs
considerably from a plant cell in terms of protein content and modification.[5] In conclusion is the Y2H
assay a very powerful method to suspect interactions but has the drawback of having to deal with a large
number of false positive and false negative results.[6] Therefore, Fu, et al. (2020) does not rely on this
technique to draw conclusions and exerted two other analyses (co-IP and BiFC). As an alternative for this
technique, a protoplast two-hybrid (P2H) assay can be used. A protoplast that resembles the plant cell of
interest is then used as the host for the two-hybrid screening. Ehlert, et al. proved this technique to be a
good alternative and tool for in vivo protein-protein interaction analysis.[7]
The results of the Y2H screening were first validated by a subcellular co-localization test. Based on the
differences between the original location of the endogenous proteins and the locations of interaction,
speculations about the influence of Cry1Ab/c were made.
To check if the proteins do indeed interact with the full-length Cry1Ab/c, a BiFC analysis is performed.
The interactions between proteins are very specific and prone to small differences in structure. As
already mentioned, the addition of an extra fragment to the interacting protein can lead to steric
hindrance and thus lead to failing interactions. To overcome this, multiple constructs must be made to
cover ideally every possible interaction manner. Fu, et al. (2020) chooses to only cover one combination
of constructs, fusing the N-terminal half to Cry1Ab/c and the C-terminal half to the interacting proteins.
To get a higher quality result, more constructs must be made.[4] To avoid this higher workload, a label
transfer screening can be used instead[8]. This uses a label transfer reagent (LTR) fused to one of the
interacting proteins. As it interacts with another protein, the LTR folds back leading to a second
interaction. As a result, the second protein now contains the photoreactive site proving the interaction
that just took place. The big advantage of using this over BiFC is that only one protein must be linked to
the LTR so that no combinations of constructs need to be made. Also, the LTR is structurally much
smaller than half of YFP, making for a lower chance of steric hindrance.
Co-IP is used as a second technique to confirm the interactions between proteins found by Y2H. As
already mentioned, is the BiFC analysis prone to spontaneous fluorescence leading to false positives,
hence the need for a second technique. The results for co-IP are largely in accordance with those from
BiFC except for one endogenous protein (CP43). According to the BiFC assay, it would interact with
Cry1Ab/c as opposed to the result of co-IP, suggesting a false positive BiFC result. The co-IP is analyzed
via SDS-PAGE, which results in Figure 1. A first critical note must be made about the lack of information
about the used extraction method. It is not indicated which buffer and protocol to use. This makes it
more difficult to get comparable results for other research. The SDS-PAGE gel itself is cut up into small
images covering only the regions where the expected bands are visible. The writers indicate the targeted
bands with a red asterisk instead of adding images of the full SDS-PAGE gels, including the used ladder
and controls. This would make for a more standardized result and unambiguous interpretation of the
data, as now there is no context to the given pictures.
To conclude, Fu, et al. (2020) uses 4 techniques to inspect the influence of Cry1Ab/c on endogenous
proteins. They use yeast two-hybrid screening to get a library of all interacting proteins and picked the
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
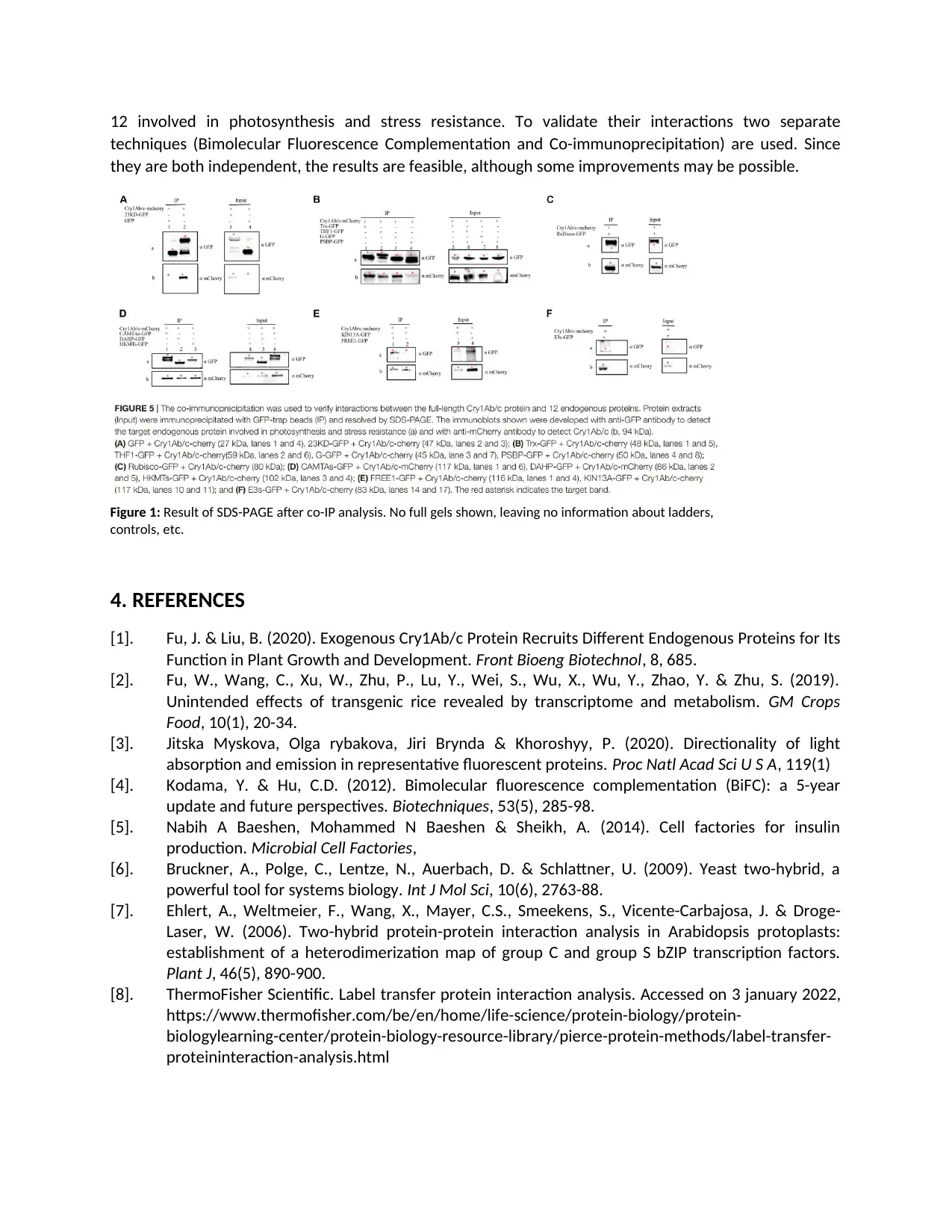
12 involved in photosynthesis and stress resistance. To validate their interactions two separate
techniques (Bimolecular Fluorescence Complementation and Co-immunoprecipitation) are used. Since
they are both independent, the results are feasible, although some improvements may be possible.
4. REFERENCES
[1]. Fu, J. & Liu, B. (2020). Exogenous Cry1Ab/c Protein Recruits Different Endogenous Proteins for Its
Function in Plant Growth and Development. Front Bioeng Biotechnol, 8, 685.
[2]. Fu, W., Wang, C., Xu, W., Zhu, P., Lu, Y., Wei, S., Wu, X., Wu, Y., Zhao, Y. & Zhu, S. (2019).
Unintended effects of transgenic rice revealed by transcriptome and metabolism. GM Crops
Food, 10(1), 20-34.
[3]. Jitska Myskova, Olga rybakova, Jiri Brynda & Khoroshyy, P. (2020). Directionality of light
absorption and emission in representative fluorescent proteins. Proc Natl Acad Sci U S A, 119(1)
[4]. Kodama, Y. & Hu, C.D. (2012). Bimolecular fluorescence complementation (BiFC): a 5-year
update and future perspectives. Biotechniques, 53(5), 285-98.
[5]. Nabih A Baeshen, Mohammed N Baeshen & Sheikh, A. (2014). Cell factories for insulin
production. Microbial Cell Factories,
[6]. Bruckner, A., Polge, C., Lentze, N., Auerbach, D. & Schlattner, U. (2009). Yeast two-hybrid, a
powerful tool for systems biology. Int J Mol Sci, 10(6), 2763-88.
[7]. Ehlert, A., Weltmeier, F., Wang, X., Mayer, C.S., Smeekens, S., Vicente-Carbajosa, J. & Droge-
Laser, W. (2006). Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts:
establishment of a heterodimerization map of group C and group S bZIP transcription factors.
Plant J, 46(5), 890-900.
[8]. ThermoFisher Scientific. Label transfer protein interaction analysis. Accessed on 3 january 2022,
https://www.thermofisher.com/be/en/home/life-science/protein-biology/protein-
biologylearning-center/protein-biology-resource-library/pierce-protein-methods/label-transfer-
proteininteraction-analysis.html
Figure 1: Result of SDS-PAGE after co-IP analysis. No full gels shown, leaving no information about ladders,
controls, etc.
techniques (Bimolecular Fluorescence Complementation and Co-immunoprecipitation) are used. Since
they are both independent, the results are feasible, although some improvements may be possible.
4. REFERENCES
[1]. Fu, J. & Liu, B. (2020). Exogenous Cry1Ab/c Protein Recruits Different Endogenous Proteins for Its
Function in Plant Growth and Development. Front Bioeng Biotechnol, 8, 685.
[2]. Fu, W., Wang, C., Xu, W., Zhu, P., Lu, Y., Wei, S., Wu, X., Wu, Y., Zhao, Y. & Zhu, S. (2019).
Unintended effects of transgenic rice revealed by transcriptome and metabolism. GM Crops
Food, 10(1), 20-34.
[3]. Jitska Myskova, Olga rybakova, Jiri Brynda & Khoroshyy, P. (2020). Directionality of light
absorption and emission in representative fluorescent proteins. Proc Natl Acad Sci U S A, 119(1)
[4]. Kodama, Y. & Hu, C.D. (2012). Bimolecular fluorescence complementation (BiFC): a 5-year
update and future perspectives. Biotechniques, 53(5), 285-98.
[5]. Nabih A Baeshen, Mohammed N Baeshen & Sheikh, A. (2014). Cell factories for insulin
production. Microbial Cell Factories,
[6]. Bruckner, A., Polge, C., Lentze, N., Auerbach, D. & Schlattner, U. (2009). Yeast two-hybrid, a
powerful tool for systems biology. Int J Mol Sci, 10(6), 2763-88.
[7]. Ehlert, A., Weltmeier, F., Wang, X., Mayer, C.S., Smeekens, S., Vicente-Carbajosa, J. & Droge-
Laser, W. (2006). Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts:
establishment of a heterodimerization map of group C and group S bZIP transcription factors.
Plant J, 46(5), 890-900.
[8]. ThermoFisher Scientific. Label transfer protein interaction analysis. Accessed on 3 january 2022,
https://www.thermofisher.com/be/en/home/life-science/protein-biology/protein-
biologylearning-center/protein-biology-resource-library/pierce-protein-methods/label-transfer-
proteininteraction-analysis.html
Figure 1: Result of SDS-PAGE after co-IP analysis. No full gels shown, leaving no information about ladders,
controls, etc.
1 out of 5
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.