Artificial Reproductive Technology in Dairy Sheep Report, AGRI4403
VerifiedAdded on 2022/12/05
|11
|2804
|308
Report
AI Summary
This report critically evaluates the application of artificial reproductive technologies (ART) in dairy sheep, focusing on the context of Clean, Green, and Ethical (CGE) management and the value of the technology to the dairy sheep industry. It explores various ART methods like in vitro fertilization, genomic selection, and SNP chips, analyzing their impact on breeding practices and production efficiency. The report discusses the benefits of these technologies, such as improved genetic traits, increased milk production, and disease resistance, while also addressing limitations like cost-effectiveness and lower linkage disequilibrium values. It examines the role of CGE management, including ethical considerations and environmental sustainability. The report highlights advancements in gender-specific breeding and the development of disease-resistant breeds. The study emphasizes the potential of genetic selection and genomic-based approaches to meet the rising global food demands and improve animal welfare. The report concludes by acknowledging the significance of ART in addressing food scarcity and promoting sustainable development in animal husbandry.

Running head: ARTIFICIAL REPRODUCTIVE TECHNOLOGY IN DAIRY SHEEP
ARTIFICIAL REPRODUCTIVE TECHNOLOGY IN DAIRY SHEEP
Name of the Student:
Name of the University:
Author Note:
ARTIFICIAL REPRODUCTIVE TECHNOLOGY IN DAIRY SHEEP
Name of the Student:
Name of the University:
Author Note:
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

1ARTIFICIAL REPRODUCTIVE TECHNOLOGY IN DAIRY SHEEP
Table of Contents
Introduction:...........................................................................................................................2
Discussion:.............................................................................................................................2
References:.............................................................................................................................8
Table of Contents
Introduction:...........................................................................................................................2
Discussion:.............................................................................................................................2
References:.............................................................................................................................8

2ARTIFICIAL REPRODUCTIVE TECHNOLOGY IN DAIRY SHEEP
Introduction:
Artificial reproductive technologies are widely used by animal breeders to obtain best
breeds with selective genetic traits that will yield higher food mass, fiber, better quality
additional products than wild type ones. Sheep have been preferentially bred for centuries to
provide as food source, wools to derive clothes and prominent succession of advantageous
genes are desirable. Epigenetic analysis and sequencing have revealed particular genes that
matches the desired phenotypes correlating with the requirement (Auvray et al. 2014).
Parental phenotypes are selected in accordance with hereditary traits via Mendelian Laws of
Inheritance. Further advancement in genetic engineering and bioinformatics have developed
innovative methods like genomic selection via SNP chips and genomic best linear unbiased
prediction (GUBP), flow cytometry based separation of sperm cells conataining X or Y
chromosome (Hansen 2013), to create best possible breeds of livestock in animal husbandry
(Baloche et al. 2014).
Discussion:
Sheep breeding is extensively performed worldwide to achieve desired progenies
maximizing the production of edible meat, wools and various dairy products. Major classical
artificial reproductive techniques include in vitro fertilization, intracytoplasmic sperm
injection, gamete intra fallopian transfer, juvenile in vitro fertilized embryo transfer (Baloche
et al. 2014).
Selective breeding with respective genotype has pooled advantageous traits in
ruminant animals. A combination of genotypic, phenotypic and pedigree based approach is
highly encouraged in breeding practice. Although, a hybrid crossing method involving
pedigree-analysis and genomic selection applied on sheep breeding have failed to satisfy
expected productive efficacy in progeny animals because of weaker carcass composition and
Introduction:
Artificial reproductive technologies are widely used by animal breeders to obtain best
breeds with selective genetic traits that will yield higher food mass, fiber, better quality
additional products than wild type ones. Sheep have been preferentially bred for centuries to
provide as food source, wools to derive clothes and prominent succession of advantageous
genes are desirable. Epigenetic analysis and sequencing have revealed particular genes that
matches the desired phenotypes correlating with the requirement (Auvray et al. 2014).
Parental phenotypes are selected in accordance with hereditary traits via Mendelian Laws of
Inheritance. Further advancement in genetic engineering and bioinformatics have developed
innovative methods like genomic selection via SNP chips and genomic best linear unbiased
prediction (GUBP), flow cytometry based separation of sperm cells conataining X or Y
chromosome (Hansen 2013), to create best possible breeds of livestock in animal husbandry
(Baloche et al. 2014).
Discussion:
Sheep breeding is extensively performed worldwide to achieve desired progenies
maximizing the production of edible meat, wools and various dairy products. Major classical
artificial reproductive techniques include in vitro fertilization, intracytoplasmic sperm
injection, gamete intra fallopian transfer, juvenile in vitro fertilized embryo transfer (Baloche
et al. 2014).
Selective breeding with respective genotype has pooled advantageous traits in
ruminant animals. A combination of genotypic, phenotypic and pedigree based approach is
highly encouraged in breeding practice. Although, a hybrid crossing method involving
pedigree-analysis and genomic selection applied on sheep breeding have failed to satisfy
expected productive efficacy in progeny animals because of weaker carcass composition and
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

3ARTIFICIAL REPRODUCTIVE TECHNOLOGY IN DAIRY SHEEP
qualitative meat, consistent with every generation (Daetwyler et al. 2012). Cost effectiveness
is also a major factor in this case, as genotyping requires a lot of money, in comparison to the
market value of the animal. Genomic selection is widely conducted in several countries like
Australia, New Zealand, UK, France (Auvray et al. 2014; Carillier et al. 2013, 2014; Mucha
et al. 2015).
Genetic selection (GS) is the future of animal husbandry to serve the rising food
requirement worldwide. A reference population consisting of wild type and a crossbred
population are required for effective outcome of genomic selection. The reference population
should have a higher number of animals compared to crossbred ones. As new traits evolved
in crossbred animals need further crossing with a reference population to exclude chances of
co-dominance, which can contribute to growth related disorders and disease susceptibility.
Further modification in genetic selection based on genomic best linear unbiased production
(GBLUP) has concluded better genomic breeding values (GEBV) than pedigree analyzed
expected breeding values (EBV).
A study by Baloche et al. (2014) has estimated that production of milk in French
Lacaune dairy sheep has increased 10-20 percent along with 14 percent increase in protein
and fat content by GBLUP mediated GEBV accuracy in GS compared to pedigree based
expected breeding value (EBV).
Genetic selection in sheep has limitations in expression of desired traits due to a lower
LD (linkage disequilibrium) value. In population genetics, linkage disequilibrium designates
the possibility of recombination of two alleles residing in close proximity. Furthermore, GS
mediated mating over 2000 reference population of sheep has optimized a 17.9% gain in
genetic traits in a year (Shumbusho et al. 2015).
qualitative meat, consistent with every generation (Daetwyler et al. 2012). Cost effectiveness
is also a major factor in this case, as genotyping requires a lot of money, in comparison to the
market value of the animal. Genomic selection is widely conducted in several countries like
Australia, New Zealand, UK, France (Auvray et al. 2014; Carillier et al. 2013, 2014; Mucha
et al. 2015).
Genetic selection (GS) is the future of animal husbandry to serve the rising food
requirement worldwide. A reference population consisting of wild type and a crossbred
population are required for effective outcome of genomic selection. The reference population
should have a higher number of animals compared to crossbred ones. As new traits evolved
in crossbred animals need further crossing with a reference population to exclude chances of
co-dominance, which can contribute to growth related disorders and disease susceptibility.
Further modification in genetic selection based on genomic best linear unbiased production
(GBLUP) has concluded better genomic breeding values (GEBV) than pedigree analyzed
expected breeding values (EBV).
A study by Baloche et al. (2014) has estimated that production of milk in French
Lacaune dairy sheep has increased 10-20 percent along with 14 percent increase in protein
and fat content by GBLUP mediated GEBV accuracy in GS compared to pedigree based
expected breeding value (EBV).
Genetic selection in sheep has limitations in expression of desired traits due to a lower
LD (linkage disequilibrium) value. In population genetics, linkage disequilibrium designates
the possibility of recombination of two alleles residing in close proximity. Furthermore, GS
mediated mating over 2000 reference population of sheep has optimized a 17.9% gain in
genetic traits in a year (Shumbusho et al. 2015).
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
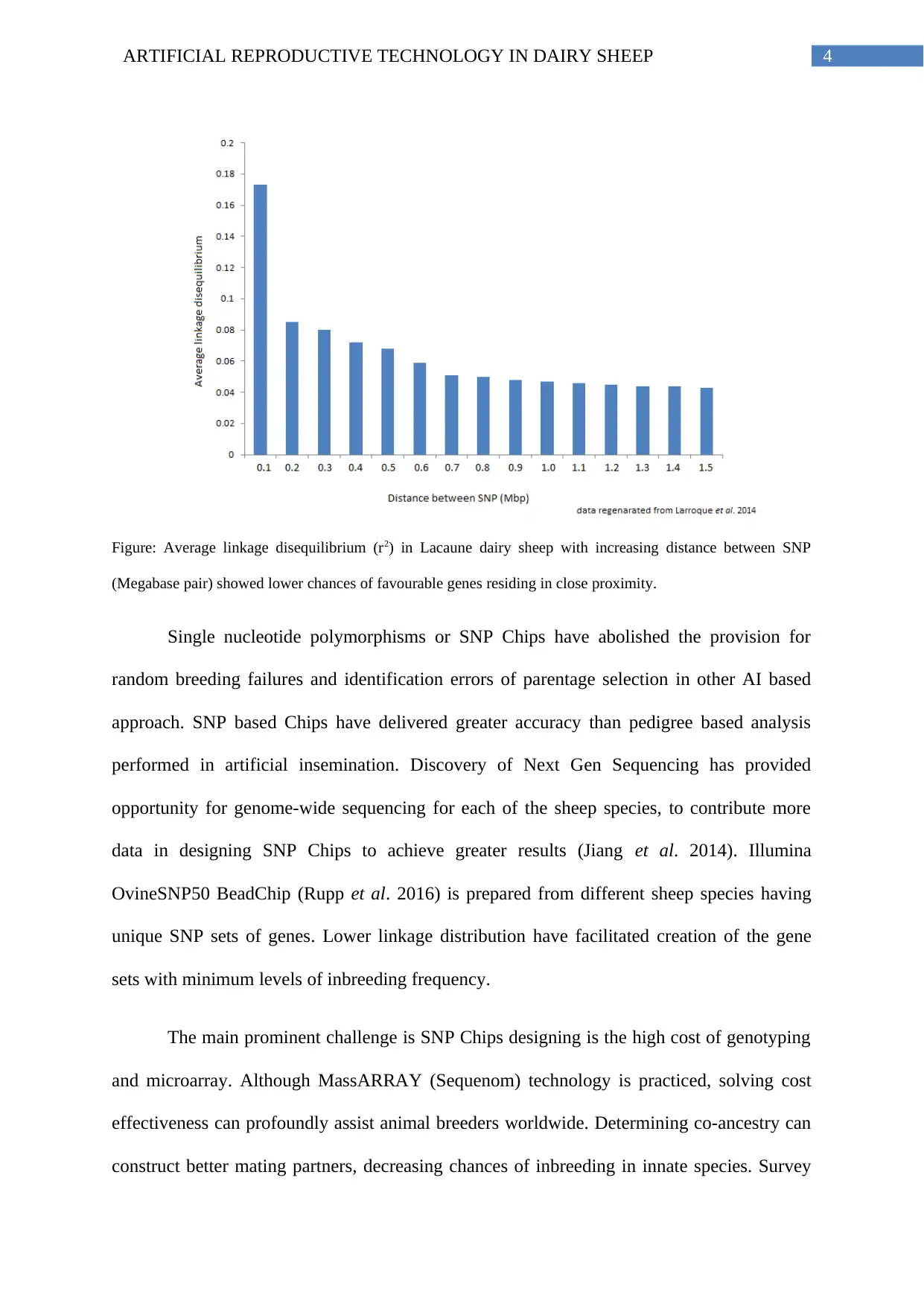
4ARTIFICIAL REPRODUCTIVE TECHNOLOGY IN DAIRY SHEEP
Figure: Average linkage disequilibrium (r2) in Lacaune dairy sheep with increasing distance between SNP
(Megabase pair) showed lower chances of favourable genes residing in close proximity.
Single nucleotide polymorphisms or SNP Chips have abolished the provision for
random breeding failures and identification errors of parentage selection in other AI based
approach. SNP based Chips have delivered greater accuracy than pedigree based analysis
performed in artificial insemination. Discovery of Next Gen Sequencing has provided
opportunity for genome-wide sequencing for each of the sheep species, to contribute more
data in designing SNP Chips to achieve greater results (Jiang et al. 2014). Illumina
OvineSNP50 BeadChip (Rupp et al. 2016) is prepared from different sheep species having
unique SNP sets of genes. Lower linkage distribution have facilitated creation of the gene
sets with minimum levels of inbreeding frequency.
The main prominent challenge is SNP Chips designing is the high cost of genotyping
and microarray. Although MassARRAY (Sequenom) technology is practiced, solving cost
effectiveness can profoundly assist animal breeders worldwide. Determining co-ancestry can
construct better mating partners, decreasing chances of inbreeding in innate species. Survey
Figure: Average linkage disequilibrium (r2) in Lacaune dairy sheep with increasing distance between SNP
(Megabase pair) showed lower chances of favourable genes residing in close proximity.
Single nucleotide polymorphisms or SNP Chips have abolished the provision for
random breeding failures and identification errors of parentage selection in other AI based
approach. SNP based Chips have delivered greater accuracy than pedigree based analysis
performed in artificial insemination. Discovery of Next Gen Sequencing has provided
opportunity for genome-wide sequencing for each of the sheep species, to contribute more
data in designing SNP Chips to achieve greater results (Jiang et al. 2014). Illumina
OvineSNP50 BeadChip (Rupp et al. 2016) is prepared from different sheep species having
unique SNP sets of genes. Lower linkage distribution have facilitated creation of the gene
sets with minimum levels of inbreeding frequency.
The main prominent challenge is SNP Chips designing is the high cost of genotyping
and microarray. Although MassARRAY (Sequenom) technology is practiced, solving cost
effectiveness can profoundly assist animal breeders worldwide. Determining co-ancestry can
construct better mating partners, decreasing chances of inbreeding in innate species. Survey

5ARTIFICIAL REPRODUCTIVE TECHNOLOGY IN DAIRY SHEEP
suggests a value of 10 to 12 dollars or less could industrialize the process in a global scale.
Alternative sequencing techniques and marker selection arrays are to be developed in future
to achieve this goal. Disease resistance is another advantage of this method, as particular
species can be intercrossed to produce a hybrid progeny. Species resistant against facial
eczema (Phua et al. 2014), parasite and fly contamination (Pickering et al. 2015) are being
developed by smaller industries. Advancement in these area can accept industrial success if
managed efficiently (Doods et al. 2015).
Controlled production of male and female progenies has been a major area of interest
in farming industry. Gender-specific breeding have improved after invention of flow
cytometry (Hansen 2013). Effeciency of producing livestock has increased at a great potential
by enabling breeders to control the proportions of animals in a specified farming area,
sustaining the ratio of male and female animals at a steady rate. Breeders can select sperm
cells containing X chromosome, extracted from a male dairy sheep to produce female
progenies. This way breeders can avoid sexual dominance in a particular population of
animals. Likewise, a breeder can also select sperm cells bearing Y chromosome to produce
male progeny of specific species to enhance the overall production of meat or wool.
However, there are few deficits in this approach. Mechanical stress during separation process
of sperm cells, laser-induced damage in flow cytometry and most of all inadequecy resulted
from in-vitro handling of the highly sensitive germ cells leads to loss of semen content of an
animal.
SNP based approach had also contributed in humanity grounds. Random mating
techniques seldom escalate into some form of disability that ends the life of the animal. The
chances of getting miscarriage are also higher in these cases. Using SNP based techniques
and genome based approach have declined the number of animal loss due to experimental
anomaly. Exposing sheep to examine disease resistance and exposure to toxic chemicals have
suggests a value of 10 to 12 dollars or less could industrialize the process in a global scale.
Alternative sequencing techniques and marker selection arrays are to be developed in future
to achieve this goal. Disease resistance is another advantage of this method, as particular
species can be intercrossed to produce a hybrid progeny. Species resistant against facial
eczema (Phua et al. 2014), parasite and fly contamination (Pickering et al. 2015) are being
developed by smaller industries. Advancement in these area can accept industrial success if
managed efficiently (Doods et al. 2015).
Controlled production of male and female progenies has been a major area of interest
in farming industry. Gender-specific breeding have improved after invention of flow
cytometry (Hansen 2013). Effeciency of producing livestock has increased at a great potential
by enabling breeders to control the proportions of animals in a specified farming area,
sustaining the ratio of male and female animals at a steady rate. Breeders can select sperm
cells containing X chromosome, extracted from a male dairy sheep to produce female
progenies. This way breeders can avoid sexual dominance in a particular population of
animals. Likewise, a breeder can also select sperm cells bearing Y chromosome to produce
male progeny of specific species to enhance the overall production of meat or wool.
However, there are few deficits in this approach. Mechanical stress during separation process
of sperm cells, laser-induced damage in flow cytometry and most of all inadequecy resulted
from in-vitro handling of the highly sensitive germ cells leads to loss of semen content of an
animal.
SNP based approach had also contributed in humanity grounds. Random mating
techniques seldom escalate into some form of disability that ends the life of the animal. The
chances of getting miscarriage are also higher in these cases. Using SNP based techniques
and genome based approach have declined the number of animal loss due to experimental
anomaly. Exposing sheep to examine disease resistance and exposure to toxic chemicals have
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

6ARTIFICIAL REPRODUCTIVE TECHNOLOGY IN DAIRY SHEEP
essentially reduced by in-vitro manipulation of the DNA markers to create a superior strain
with higher fidelity. Several genes have been augmented in sheep breeding to reach better
progeny outcomes like- GDF8 gene for muscle development (Clop et al. 2006), BCO2 gene
to produce higher amount of yellow fat (Vage and Boman 2010), Bmp15 Gdf9 and FecL
genes for to achieve effective proliferation (Demars et al. 2013; Martin et al. 2014), Prp gene
to build resistance against scrapie disease (Elsen et al. 1999). Current and further invention in
these major breakthroughs are encouraged and supported by International Goat Genomics
Consortia (IGGC).
Previously it was believed that tying up right testicle of a sheep would produce ewe
(female sheep), and if the left is tied, it will generate a male sheep. This strategy was widely
used upto early 1900’s to attain required progenies. The amount of agony that a sheep had to
experience in this process is unfathomably painful. Invention of flow cytometry has abolished
the chances of this inhuman practices, except poorly educated areas, as isolating semen
sample from sheep and differentiating between X and Y chromosome spermatozoa under
laboratory conditions are far more ethical and humane practice compared to the conventional
strategy mentioned above.
The concept of clean, green and ethical management was first propounded in the year
2004 and it intended to provide an insight to the farmers to effectively address societal
demands and improve the diary management process. The technologies are mainly based on
the inclusion of tactics such as minimal drug use, minimising enviornmental footprint so as to
maximise animal welfare. A number of genetic studies and research in this area would
ultimately lead to the development of effective technologies that would improve the animal
health outcome and positively enhance the quality of production and would serve as a means
of sustainable development. However, it can be stated that the major limitations of the Clean
and Green technology framework comprise of elements that include developing drug-free
essentially reduced by in-vitro manipulation of the DNA markers to create a superior strain
with higher fidelity. Several genes have been augmented in sheep breeding to reach better
progeny outcomes like- GDF8 gene for muscle development (Clop et al. 2006), BCO2 gene
to produce higher amount of yellow fat (Vage and Boman 2010), Bmp15 Gdf9 and FecL
genes for to achieve effective proliferation (Demars et al. 2013; Martin et al. 2014), Prp gene
to build resistance against scrapie disease (Elsen et al. 1999). Current and further invention in
these major breakthroughs are encouraged and supported by International Goat Genomics
Consortia (IGGC).
Previously it was believed that tying up right testicle of a sheep would produce ewe
(female sheep), and if the left is tied, it will generate a male sheep. This strategy was widely
used upto early 1900’s to attain required progenies. The amount of agony that a sheep had to
experience in this process is unfathomably painful. Invention of flow cytometry has abolished
the chances of this inhuman practices, except poorly educated areas, as isolating semen
sample from sheep and differentiating between X and Y chromosome spermatozoa under
laboratory conditions are far more ethical and humane practice compared to the conventional
strategy mentioned above.
The concept of clean, green and ethical management was first propounded in the year
2004 and it intended to provide an insight to the farmers to effectively address societal
demands and improve the diary management process. The technologies are mainly based on
the inclusion of tactics such as minimal drug use, minimising enviornmental footprint so as to
maximise animal welfare. A number of genetic studies and research in this area would
ultimately lead to the development of effective technologies that would improve the animal
health outcome and positively enhance the quality of production and would serve as a means
of sustainable development. However, it can be stated that the major limitations of the Clean
and Green technology framework comprise of elements that include developing drug-free
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

7ARTIFICIAL REPRODUCTIVE TECHNOLOGY IN DAIRY SHEEP
control of reproduction, fertility, fecundity, colostrum production, weaner mortality and
mother-young bonding. Extensive research in these areas and limiting the use of chemicals
and artificial hormones can help in implementing CHE technology and maintain high
standard of animal welfare.
Conclusion:
In this post modernization era, food scarcity and mean nutrition rate in human beings
has been a utmost crisis across global countries and populations. Finding alternative food
source and enhancement of the existing one are two realistic possibilities that urge focus to
address these parameters. Although, opportunities for artificial insemination (AI) is limited,
and certain conditions of pedigree-based approach are unsure and error prone, exploring
genetics and molecular biology based themes are genuinely intriguing to assess efficiency in
providing better results in animal husbandry and inter species breeding.
control of reproduction, fertility, fecundity, colostrum production, weaner mortality and
mother-young bonding. Extensive research in these areas and limiting the use of chemicals
and artificial hormones can help in implementing CHE technology and maintain high
standard of animal welfare.
Conclusion:
In this post modernization era, food scarcity and mean nutrition rate in human beings
has been a utmost crisis across global countries and populations. Finding alternative food
source and enhancement of the existing one are two realistic possibilities that urge focus to
address these parameters. Although, opportunities for artificial insemination (AI) is limited,
and certain conditions of pedigree-based approach are unsure and error prone, exploring
genetics and molecular biology based themes are genuinely intriguing to assess efficiency in
providing better results in animal husbandry and inter species breeding.

8ARTIFICIAL REPRODUCTIVE TECHNOLOGY IN DAIRY SHEEP
References:
Auvray, B., McEwan, J.C., Newman, S.A., Lee, M. and Dodds, K.G., 2014. Genomic
prediction of breeding values in the New Zealand sheep industry using a 50K SNP
chip. Journal of animal science, 92(10), pp.4375-4389.
Baloche, G., Legarra, A., Sallé, G., Larroque, H., Astruc, J.M., Robert-Granié, C. and
Barillet, F., 2014. Assessment of accuracy of genomic prediction for French Lacaune dairy
sheep. Journal of dairy science, 97(2), pp.1107-1116.
Carillier, C., Larroque, H. and Robert-Granié, C., 2014. Comparison of joint versus purebred
genomic evaluation in the French multi-breed dairy goat population. Genetics Selection
Evolution, 46(1), p.67.
Carillier, C., Larroque, H., Palhiere, I., Clément, V., Rupp, R. and Robert-Granié, C., 2013. A
first step toward genomic selection in the multi-breed French dairy goat population. Journal
of dairy science, 96(11), pp.7294-7305.
Clop, A., Marcq, F., Takeda, H., Pirottin, D., Tordoir, X., Bibé, B., Bouix, J., Caiment, F.,
Elsen, J.M., Eychenne, F. and Larzul, C., 2006. A mutation creating a potential illegitimate
microRNA target site in the myostatin gene affects muscularity in sheep. Nature
genetics, 38(7), p.813.
Daetwyler, H.D., Swan, A.A., van der Werf, J.H. and Hayes, B.J., 2012. Accuracy of
pedigree and genomic predictions of carcass and novel meat quality traits in multi-breed
sheep data assessed by cross-validation. Genetics Selection Evolution, 44(1), p.33.
Demars, J., Fabre, S., Sarry, J., Rossetti, R., Gilbert, H., Persani, L., Tosser-Klopp, G.,
Mulsant, P., Nowak, Z., Drobik, W. and Martyniuk, E., 2013. Genome-wide association
References:
Auvray, B., McEwan, J.C., Newman, S.A., Lee, M. and Dodds, K.G., 2014. Genomic
prediction of breeding values in the New Zealand sheep industry using a 50K SNP
chip. Journal of animal science, 92(10), pp.4375-4389.
Baloche, G., Legarra, A., Sallé, G., Larroque, H., Astruc, J.M., Robert-Granié, C. and
Barillet, F., 2014. Assessment of accuracy of genomic prediction for French Lacaune dairy
sheep. Journal of dairy science, 97(2), pp.1107-1116.
Carillier, C., Larroque, H. and Robert-Granié, C., 2014. Comparison of joint versus purebred
genomic evaluation in the French multi-breed dairy goat population. Genetics Selection
Evolution, 46(1), p.67.
Carillier, C., Larroque, H., Palhiere, I., Clément, V., Rupp, R. and Robert-Granié, C., 2013. A
first step toward genomic selection in the multi-breed French dairy goat population. Journal
of dairy science, 96(11), pp.7294-7305.
Clop, A., Marcq, F., Takeda, H., Pirottin, D., Tordoir, X., Bibé, B., Bouix, J., Caiment, F.,
Elsen, J.M., Eychenne, F. and Larzul, C., 2006. A mutation creating a potential illegitimate
microRNA target site in the myostatin gene affects muscularity in sheep. Nature
genetics, 38(7), p.813.
Daetwyler, H.D., Swan, A.A., van der Werf, J.H. and Hayes, B.J., 2012. Accuracy of
pedigree and genomic predictions of carcass and novel meat quality traits in multi-breed
sheep data assessed by cross-validation. Genetics Selection Evolution, 44(1), p.33.
Demars, J., Fabre, S., Sarry, J., Rossetti, R., Gilbert, H., Persani, L., Tosser-Klopp, G.,
Mulsant, P., Nowak, Z., Drobik, W. and Martyniuk, E., 2013. Genome-wide association
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

9ARTIFICIAL REPRODUCTIVE TECHNOLOGY IN DAIRY SHEEP
studies identify two novel BMP15 mutations responsible for an atypical hyperprolificacy
phenotype in sheep. PLoS genetics, 9(4), p.e1003482.
Dodds, K.G., McEwan, J.C., Brauning, R., Anderson, R.M., Stijn, T.C., Kristjánsson, T. and
Clarke, S.M., 2015. Construction of relatedness matrices using genotyping-by-sequencing
data. BMC genomics, 16(1), p.1047.
Elsen, J.M., Amigues, Y., Schelcher, F., Ducrocq, V., Andreoletti, O., Eychenne, F., Khang,
J.T., Poivey, J.P., Lantier, F. and Laplanche, J.L., 1999. Genetic susceptibility and
transmission factors in scrapie: detailed analysis of an epidemic in a closed flock of
Romanov. Archives of virology, 144(3), pp.431-445.
Hansen, P.J., 2014. Current and future assisted reproductive technologies for mammalian
farm animals. In Current and future reproductive technologies and world food production
(pp. 1-22). Springer, New York, NY.
Jiang, Y., Xie, M., Chen, W., Talbot, R., Maddox, J.F., Faraut, T., Wu, C., Muzny, D.M., Li,
Y., Zhang, W. and Stanton, J.A., 2014. The sheep genome illuminates biology of the rumen
and lipid metabolism. Science, 344(6188), pp.1168-1173.
Larroque, H., Barillet, F., Baloche, G., Astruc, J.M., Buisson, D., Shumbusho, F., Clément,
V., Lagriffoul, G., Palhiere, I., Rupp, R. and Carillier, C., 2014, August. Toward genomic
breeding programs in French dairy sheep and goats. In Vancouver, Canada: 10th World
Congress on Genetics Applied to Livestock Production.
Martin, G.B. and Ferasyi, T.R., 2016. 1. Clean, Green, Ethical (CGE) Management: What
Research Do We Really Need?. The International Journal of Tropical Veterinary and
Biomedical Research, 1(1), pp.1-8.
studies identify two novel BMP15 mutations responsible for an atypical hyperprolificacy
phenotype in sheep. PLoS genetics, 9(4), p.e1003482.
Dodds, K.G., McEwan, J.C., Brauning, R., Anderson, R.M., Stijn, T.C., Kristjánsson, T. and
Clarke, S.M., 2015. Construction of relatedness matrices using genotyping-by-sequencing
data. BMC genomics, 16(1), p.1047.
Elsen, J.M., Amigues, Y., Schelcher, F., Ducrocq, V., Andreoletti, O., Eychenne, F., Khang,
J.T., Poivey, J.P., Lantier, F. and Laplanche, J.L., 1999. Genetic susceptibility and
transmission factors in scrapie: detailed analysis of an epidemic in a closed flock of
Romanov. Archives of virology, 144(3), pp.431-445.
Hansen, P.J., 2014. Current and future assisted reproductive technologies for mammalian
farm animals. In Current and future reproductive technologies and world food production
(pp. 1-22). Springer, New York, NY.
Jiang, Y., Xie, M., Chen, W., Talbot, R., Maddox, J.F., Faraut, T., Wu, C., Muzny, D.M., Li,
Y., Zhang, W. and Stanton, J.A., 2014. The sheep genome illuminates biology of the rumen
and lipid metabolism. Science, 344(6188), pp.1168-1173.
Larroque, H., Barillet, F., Baloche, G., Astruc, J.M., Buisson, D., Shumbusho, F., Clément,
V., Lagriffoul, G., Palhiere, I., Rupp, R. and Carillier, C., 2014, August. Toward genomic
breeding programs in French dairy sheep and goats. In Vancouver, Canada: 10th World
Congress on Genetics Applied to Livestock Production.
Martin, G.B. and Ferasyi, T.R., 2016. 1. Clean, Green, Ethical (CGE) Management: What
Research Do We Really Need?. The International Journal of Tropical Veterinary and
Biomedical Research, 1(1), pp.1-8.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

10ARTIFICIAL REPRODUCTIVE TECHNOLOGY IN DAIRY SHEEP
Martin, P., Raoul, J. and Bodin, L., 2014. Effects of the FecL major gene in the Lacaune meat
sheep population. Genetics Selection Evolution, 46(1), p.48.
Phua, S.H., Hyndman, D.L., Baird, H.J., Auvray, B., McEwan, J.C., Lee, M.A. and Dodds,
K.G., 2014. Towards genomic selection for facial eczema disease tolerance in the New
Zealand sheep industry. Animal genetics, 45(4), pp.559-564.
Pickering, N.K., Oddy, V.H., Basarab, J., Cammack, K., Hayes, B., Hegarty, R.S., Lassen, J.,
McEwan, J.C., Miller, S., Pinares-Patiño, C.S. and De Haas, Y., 2015. Animal board invited
review: genetic possibilities to reduce enteric methane emissions from
ruminants. Animal, 9(9), pp.1431-1440.
Rupp, R., Mucha, S., Larroque, H., McEwan, J. and Conington, J., 2016. Genomic
application in sheep and goat breeding. Animal Frontiers, 6(1), pp.39-44.
Shumbusho, F., Raoul, J., Astruc, J.M., Palhiere, I., Lemarié, S., Fugeray-Scarbel, A. and
Elsen, J.M., 2016. Economic evaluation of genomic selection in small ruminants: a sheep
meat breeding program. Animal, 10(6), pp.1033-1041.
Våge, D.I. and Boman, I.A., 2010. A nonsense mutation in the beta-carotene oxygenase 2
(BCO2) gene is tightly associated with accumulation of carotenoids in adipose tissue in sheep
(Ovis aries). BMC genetics, 11(1), p.10.
Martin, P., Raoul, J. and Bodin, L., 2014. Effects of the FecL major gene in the Lacaune meat
sheep population. Genetics Selection Evolution, 46(1), p.48.
Phua, S.H., Hyndman, D.L., Baird, H.J., Auvray, B., McEwan, J.C., Lee, M.A. and Dodds,
K.G., 2014. Towards genomic selection for facial eczema disease tolerance in the New
Zealand sheep industry. Animal genetics, 45(4), pp.559-564.
Pickering, N.K., Oddy, V.H., Basarab, J., Cammack, K., Hayes, B., Hegarty, R.S., Lassen, J.,
McEwan, J.C., Miller, S., Pinares-Patiño, C.S. and De Haas, Y., 2015. Animal board invited
review: genetic possibilities to reduce enteric methane emissions from
ruminants. Animal, 9(9), pp.1431-1440.
Rupp, R., Mucha, S., Larroque, H., McEwan, J. and Conington, J., 2016. Genomic
application in sheep and goat breeding. Animal Frontiers, 6(1), pp.39-44.
Shumbusho, F., Raoul, J., Astruc, J.M., Palhiere, I., Lemarié, S., Fugeray-Scarbel, A. and
Elsen, J.M., 2016. Economic evaluation of genomic selection in small ruminants: a sheep
meat breeding program. Animal, 10(6), pp.1033-1041.
Våge, D.I. and Boman, I.A., 2010. A nonsense mutation in the beta-carotene oxygenase 2
(BCO2) gene is tightly associated with accumulation of carotenoids in adipose tissue in sheep
(Ovis aries). BMC genetics, 11(1), p.10.
1 out of 11
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2025 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.