Enzyme Kinetics and Acid Phosphatase: Experimental Studies
VerifiedAdded on 2023/05/31
|12
|1884
|76
AI Summary
This article discusses experimental studies on enzyme kinetics and acid phosphatase, including the properties of Benzonase endonuclease and acid phosphatase from wheat germ. The article also covers the use of para-nitrophenylphosphate (p-NPP) as a substrate for measuring the kinetic of acid phosphatase.
Contribute Materials
Your contribution can guide someone’s learning journey. Share your
documents today.

Enzyme kinetics1
ENZYME KINETICS
Name:
Department:
School:
Date:
ENZYME KINETICS
Name:
Department:
School:
Date:
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

Enzyme kinetics 2
Paper 1
Introduction
Benzonase is a genetically engineered endonuclease from the rod-shaped gram-negative bacteria
serratia marcescens. The protein is a dimer of 30kDa sub divisions, which degrades RNA and
DNA. It is active over a broad range of situation. Benzonase fully digests nucleic acids to trivial
3-5bp oligonucleotides. This is ideal for eradication of nucleic acid from recombinant proteins
and for use where full digestion of nucleic acids is desirable. Pre-treatment of a protein samples
advances its appearances on a western blot by removing any bound nucleic acid. U.S. FDA
guidelines for the manufacture of the recombinant biological for therapeutic use demands that
nucleic acid contamination ought to be restricted to 100pg per dose. Benzonase, when utilised
under suitable reaction states, will degrade all nucleic acid sets down to oligonucleotides of
about 3-5 base pairs in length, which is considerably below the hybridization limit. The
capability of Benzonase to quickly hydrolyze nucleic acids makes the enzyme appropriate for
minimizing cell lysate viscosity both in the manufacturing plant and research laboratory.
Benzonase also allow reduction of processing time, increase the yield of protein products,
improve the separation of supernatants and pellets in centrifugation, facilitate the solutions
filtration particularly the ultrafiltration, increase the efficiency of chromatographic purification
phases and finally boost the productivity of cross-flow microfiltration steps. It is well known
that nucleic acid may adhere to cell-derived bits such as inclusion bodies or viruses. This
adhesion many interfere with separation due to agglomeration, change in particle size or charge,
resulting in a minimised product yield. Thus, Benzonase is well fitted for lessening the nucleic
acid load during purification, thus reducing interferences and advancing both the purity and yield
of the end products.
Paper 1
Introduction
Benzonase is a genetically engineered endonuclease from the rod-shaped gram-negative bacteria
serratia marcescens. The protein is a dimer of 30kDa sub divisions, which degrades RNA and
DNA. It is active over a broad range of situation. Benzonase fully digests nucleic acids to trivial
3-5bp oligonucleotides. This is ideal for eradication of nucleic acid from recombinant proteins
and for use where full digestion of nucleic acids is desirable. Pre-treatment of a protein samples
advances its appearances on a western blot by removing any bound nucleic acid. U.S. FDA
guidelines for the manufacture of the recombinant biological for therapeutic use demands that
nucleic acid contamination ought to be restricted to 100pg per dose. Benzonase, when utilised
under suitable reaction states, will degrade all nucleic acid sets down to oligonucleotides of
about 3-5 base pairs in length, which is considerably below the hybridization limit. The
capability of Benzonase to quickly hydrolyze nucleic acids makes the enzyme appropriate for
minimizing cell lysate viscosity both in the manufacturing plant and research laboratory.
Benzonase also allow reduction of processing time, increase the yield of protein products,
improve the separation of supernatants and pellets in centrifugation, facilitate the solutions
filtration particularly the ultrafiltration, increase the efficiency of chromatographic purification
phases and finally boost the productivity of cross-flow microfiltration steps. It is well known
that nucleic acid may adhere to cell-derived bits such as inclusion bodies or viruses. This
adhesion many interfere with separation due to agglomeration, change in particle size or charge,
resulting in a minimised product yield. Thus, Benzonase is well fitted for lessening the nucleic
acid load during purification, thus reducing interferences and advancing both the purity and yield
of the end products.

Enzyme kinetics 3
Benzonase endonuclease is a protein comprising of two subunits with a molecular weight of
approximately 30kD each. The protein has an isoelectric point (pi) at pH 6.85, and it functional
is between 6 and 10, and from o0c to above 420c Mg 2+ (1-2mM) is needed for the enzyme
activity. Benzonase endonuclease acts as an endonuclease that degrades both the RNA and DNA,
whether single-stranded, double-stranded, circular, linear, or supercoiled. Benzonase hydrolyses
internal phosphodiester bonds present between the nucleotides. Upon full digestion, all free
nucleic acids present in solution are minimised to 5-monophosphate-terminated oligonucleotides
which are three to five bases in length.
Methods
Bacteria transformation is the routine of introducing foreign plasmid DNA into bacteria, with the
intention of expressing a protein for purification or propagating the plasmid in bacteria then
purifying the plasmid for applications. Using DH5a E. coli cells which was made chemically
competent by addition of salt solution to permit the bacteria to open up their cell wall pores to let
the plasmid come in.
Following transformation he bacteria were plated on LB agars containing Kanamycin so that the
following day could form single discreet colonies, all of which are the outcomes of a single
bacteria taking up the plasmid. The colony picking process began and the picked colony were
inoculated into LB liquid broth comprising the kanamycin overnight, so as to permit the bacteria
to grow, continually replicating the plasmid and offering with a reasonable plasmid yield of
DNA after bacterial lysis and DNA purification.
The important of miniprep skin kit is to prepare a sample of purified DNA for subsequent
digestion. The miniprep kit reagent contained P2 buffer comprising NAOH and sodium dodecyl
Benzonase endonuclease is a protein comprising of two subunits with a molecular weight of
approximately 30kD each. The protein has an isoelectric point (pi) at pH 6.85, and it functional
is between 6 and 10, and from o0c to above 420c Mg 2+ (1-2mM) is needed for the enzyme
activity. Benzonase endonuclease acts as an endonuclease that degrades both the RNA and DNA,
whether single-stranded, double-stranded, circular, linear, or supercoiled. Benzonase hydrolyses
internal phosphodiester bonds present between the nucleotides. Upon full digestion, all free
nucleic acids present in solution are minimised to 5-monophosphate-terminated oligonucleotides
which are three to five bases in length.
Methods
Bacteria transformation is the routine of introducing foreign plasmid DNA into bacteria, with the
intention of expressing a protein for purification or propagating the plasmid in bacteria then
purifying the plasmid for applications. Using DH5a E. coli cells which was made chemically
competent by addition of salt solution to permit the bacteria to open up their cell wall pores to let
the plasmid come in.
Following transformation he bacteria were plated on LB agars containing Kanamycin so that the
following day could form single discreet colonies, all of which are the outcomes of a single
bacteria taking up the plasmid. The colony picking process began and the picked colony were
inoculated into LB liquid broth comprising the kanamycin overnight, so as to permit the bacteria
to grow, continually replicating the plasmid and offering with a reasonable plasmid yield of
DNA after bacterial lysis and DNA purification.
The important of miniprep skin kit is to prepare a sample of purified DNA for subsequent
digestion. The miniprep kit reagent contained P2 buffer comprising NAOH and sodium dodecyl

Enzyme kinetics 4
sulphate which could bust open bacteria in second, buffer P3 and buffer PB. Mini-prep (small-
scale plasmid preparation) was used to purify plasmid DNA using a spin column-based kit from
Qiagenn. The column comprised a silica membrane that binds up to 0ug DNA but only in the
present of high concentration of chaotropic salt.
Then the Benzonase digestion took place where the enzyme completely digested the nucleic
acids to form small 3-5bp oligonucleotides. The method is idea for the removal of nucleic acids
from the recombinant proteins and for use where complete digestion of nucleic acids is desirable.
It is worth noting that pre-treatment of a protein samples improves its outlook on a western blot,
by eliminating any bound nucleic acids. The experiment digested the plasmid DNA created
during the practical using Benzonase, differing metal ion strength, and pH and temperature to
determine approximate optima. The DNA was run in an agarose to assess the extent of its
degradation. After the gel was run, it was analyzed on David Pryce’s gel imager on level 3.
Results
Figure 1: Uncut/pH 4/pH 7/ pH 10
sulphate which could bust open bacteria in second, buffer P3 and buffer PB. Mini-prep (small-
scale plasmid preparation) was used to purify plasmid DNA using a spin column-based kit from
Qiagenn. The column comprised a silica membrane that binds up to 0ug DNA but only in the
present of high concentration of chaotropic salt.
Then the Benzonase digestion took place where the enzyme completely digested the nucleic
acids to form small 3-5bp oligonucleotides. The method is idea for the removal of nucleic acids
from the recombinant proteins and for use where complete digestion of nucleic acids is desirable.
It is worth noting that pre-treatment of a protein samples improves its outlook on a western blot,
by eliminating any bound nucleic acids. The experiment digested the plasmid DNA created
during the practical using Benzonase, differing metal ion strength, and pH and temperature to
determine approximate optima. The DNA was run in an agarose to assess the extent of its
degradation. After the gel was run, it was analyzed on David Pryce’s gel imager on level 3.
Results
Figure 1: Uncut/pH 4/pH 7/ pH 10
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.
Enzyme kinetics 5
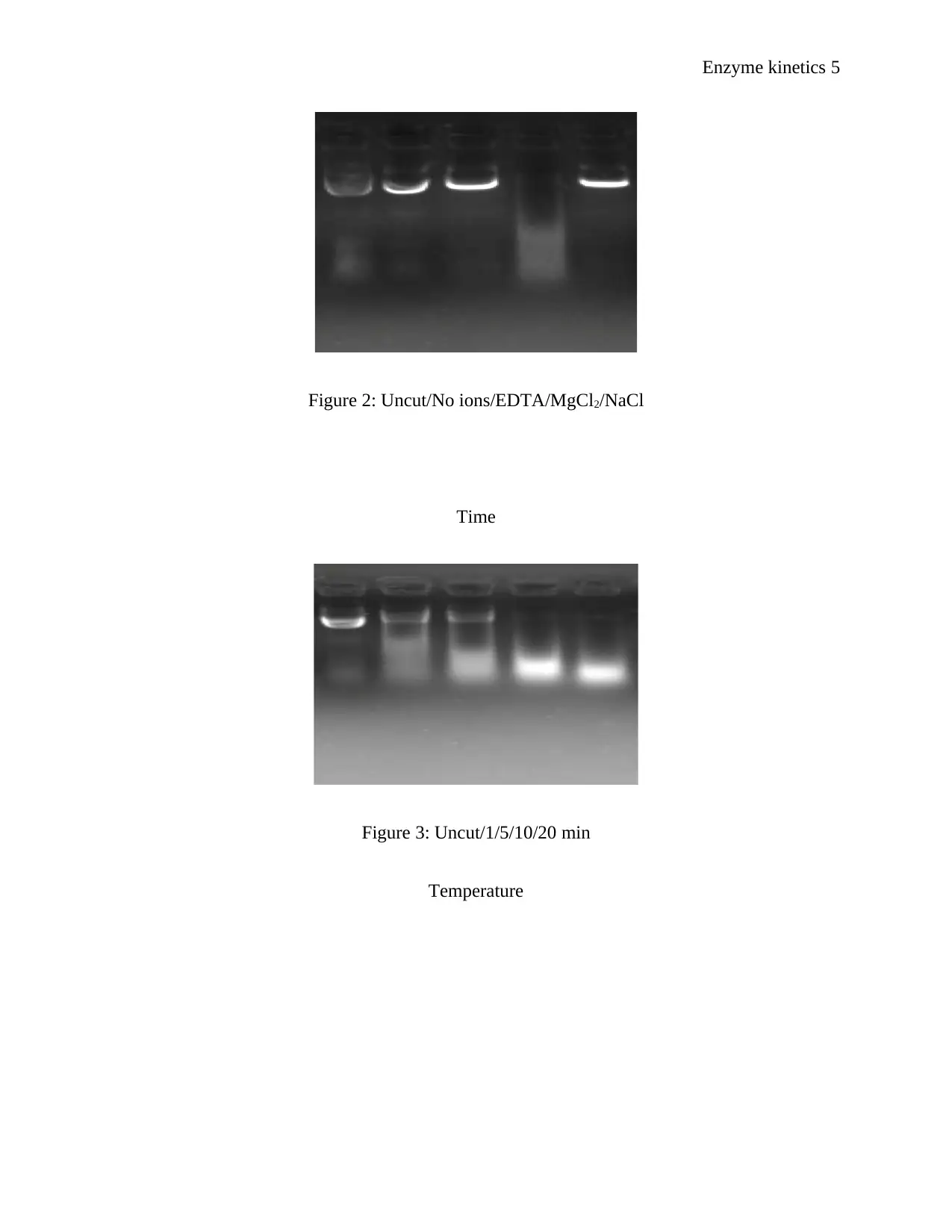
Figure 2: Uncut/No ions/EDTA/MgCl2/NaCl
Time
Figure 3: Uncut/1/5/10/20 min
Temperature
Figure 2: Uncut/No ions/EDTA/MgCl2/NaCl
Time
Figure 3: Uncut/1/5/10/20 min
Temperature

Enzyme kinetics 6
Figure 4: 0, Room Temp, 37 and 65 degrees Celsius
Discussion
Figure 4: 0, Room Temp, 37 and 65 degrees Celsius
Discussion

Enzyme kinetics 7
Paper 2
Acid phosphate kinetics
Acid phosphatase is an enzyme with phosphomonoesterase activity, catalyzing the hydrolysis of
phosphate groups from biological macromolecules. The endocytosis of cell surface
macromolecules comprises trafficking over endosomes prior to endosome-lysosome fusion.
Since endosomes become progressively acidified during the function, the enzyme functions best
at relatively low pH. Several substances have been developed that are changed to a chromophore
upon hydrolysis of the phosphate moiety. Thus, there are good substrates for measuring the
kinetic of acid phosphatase. I this experiment para-nitrophenylphosphate (p-NPP) will be used,
which by itself have no absorbance at the visible spectrum. But, one of its hydrolysis products of
p-NPP is 4-nitrophenol, which under alkaline states is transformed to a p-nitrophenolate ion,
which is yellow and can thus be detected using a spectrophotometer. This absorbance offers a
means to evaluate the strength of the reaction products, and therefore the rate of the reaction.
Thus, the phosphatase activity can be checked by determining the increase in absorbance at
405nm that results from the 4-nitropheonlate ion release.
During this trial, investigation of properties of an acid phosphatase from wheat germ was done.
Acid phosphatases have an optimal activity at pH values from 4 to 6 and are distinct from
alkaline phosphatases with optimal activity in the range of 4-9. Therefore, the kinetic
experiment investigates the effect of reaction conditions on the kinetic properties of wheat germ
acid phosphatase. Kinetics concerns the research of reaction rates and the conditions that affect
the reaction rates. One of the key concerns of kinetics deals with the examination of the variation
in the rates of the reaction as functions of the substrate or reactant concentrations. Therefore,
Paper 2
Acid phosphate kinetics
Acid phosphatase is an enzyme with phosphomonoesterase activity, catalyzing the hydrolysis of
phosphate groups from biological macromolecules. The endocytosis of cell surface
macromolecules comprises trafficking over endosomes prior to endosome-lysosome fusion.
Since endosomes become progressively acidified during the function, the enzyme functions best
at relatively low pH. Several substances have been developed that are changed to a chromophore
upon hydrolysis of the phosphate moiety. Thus, there are good substrates for measuring the
kinetic of acid phosphatase. I this experiment para-nitrophenylphosphate (p-NPP) will be used,
which by itself have no absorbance at the visible spectrum. But, one of its hydrolysis products of
p-NPP is 4-nitrophenol, which under alkaline states is transformed to a p-nitrophenolate ion,
which is yellow and can thus be detected using a spectrophotometer. This absorbance offers a
means to evaluate the strength of the reaction products, and therefore the rate of the reaction.
Thus, the phosphatase activity can be checked by determining the increase in absorbance at
405nm that results from the 4-nitropheonlate ion release.
During this trial, investigation of properties of an acid phosphatase from wheat germ was done.
Acid phosphatases have an optimal activity at pH values from 4 to 6 and are distinct from
alkaline phosphatases with optimal activity in the range of 4-9. Therefore, the kinetic
experiment investigates the effect of reaction conditions on the kinetic properties of wheat germ
acid phosphatase. Kinetics concerns the research of reaction rates and the conditions that affect
the reaction rates. One of the key concerns of kinetics deals with the examination of the variation
in the rates of the reaction as functions of the substrate or reactant concentrations. Therefore,
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
Enzyme kinetics 8
such measurement can lead to the rate constant calculation, and where suitable, equilibrium
constant. According to the Michaelis-Menten study, there several assumptions; substrates and
enzyme ought to physically combine for the catalysis to happen; the enzyme surface has only a
limited number of active sites where the catalysis actually takes place; finally, the revisable
binding of the enzyme and substrate happens much more rapidly than subsequent bond breaking
phases.
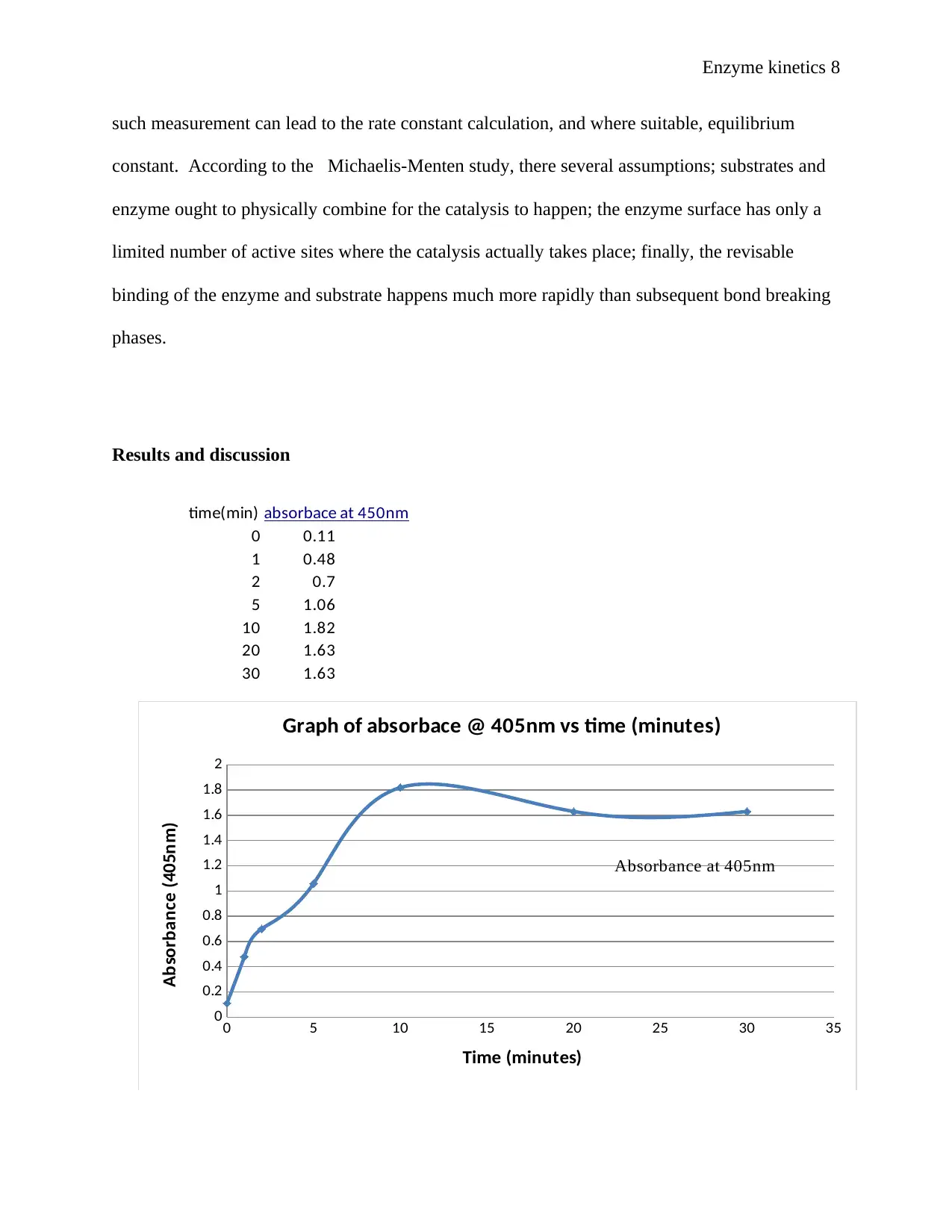
Results and discussion
time(min)
0 0.11
1 0.48
2 0.7
5 1.06
10 1.82
20 1.63
30 1.63
absorbace at 450nm
0 5 10 15 20 25 30 35
0
0.2
0.4
0.6
0.8
1
1.2
1.4
1.6
1.8
2
Graph of absorbace @ 405nm vs time (minutes)
Time (minutes)
Absorbance (405nm)
Absorbance at 405nm
such measurement can lead to the rate constant calculation, and where suitable, equilibrium
constant. According to the Michaelis-Menten study, there several assumptions; substrates and
enzyme ought to physically combine for the catalysis to happen; the enzyme surface has only a
limited number of active sites where the catalysis actually takes place; finally, the revisable
binding of the enzyme and substrate happens much more rapidly than subsequent bond breaking
phases.
Results and discussion
time(min)
0 0.11
1 0.48
2 0.7
5 1.06
10 1.82
20 1.63
30 1.63
absorbace at 450nm
0 5 10 15 20 25 30 35
0
0.2
0.4
0.6
0.8
1
1.2
1.4
1.6
1.8
2
Graph of absorbace @ 405nm vs time (minutes)
Time (minutes)
Absorbance (405nm)
Absorbance at 405nm
Enzyme kinetics 9
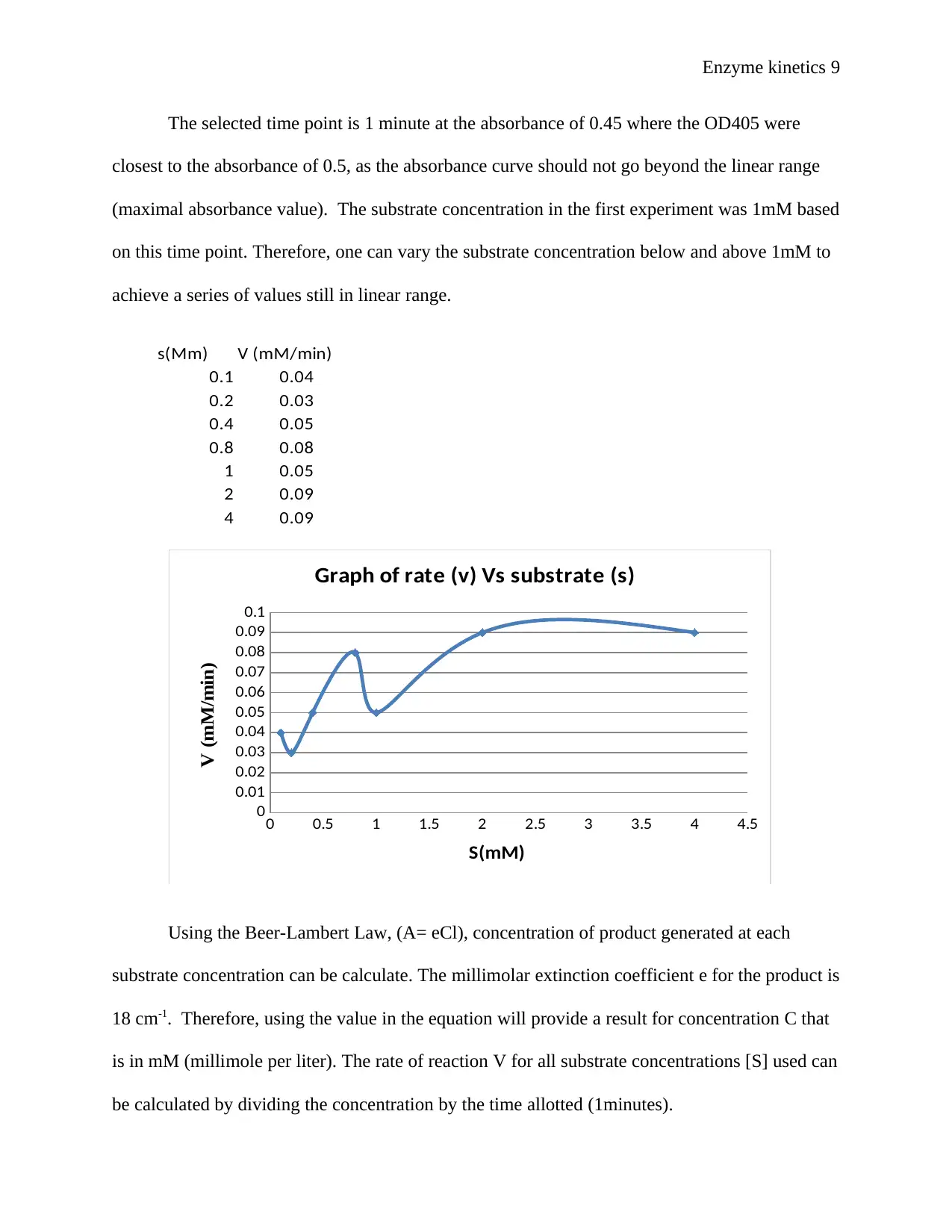
The selected time point is 1 minute at the absorbance of 0.45 where the OD405 were
closest to the absorbance of 0.5, as the absorbance curve should not go beyond the linear range
(maximal absorbance value). The substrate concentration in the first experiment was 1mM based
on this time point. Therefore, one can vary the substrate concentration below and above 1mM to
achieve a series of values still in linear range.
s(Mm) V (mM/min)
0.1 0.04
0.2 0.03
0.4 0.05
0.8 0.08
1 0.05
2 0.09
4 0.09
0 0.5 1 1.5 2 2.5 3 3.5 4 4.5
0
0.01
0.02
0.03
0.04
0.05
0.06
0.07
0.08
0.09
0.1
Graph of rate (v) Vs substrate (s)
S(mM)
V (mM/min)
Using the Beer-Lambert Law, (A= eCl), concentration of product generated at each
substrate concentration can be calculate. The millimolar extinction coefficient e for the product is
18 cm-1. Therefore, using the value in the equation will provide a result for concentration C that
is in mM (millimole per liter). The rate of reaction V for all substrate concentrations [S] used can
be calculated by dividing the concentration by the time allotted (1minutes).
The selected time point is 1 minute at the absorbance of 0.45 where the OD405 were
closest to the absorbance of 0.5, as the absorbance curve should not go beyond the linear range
(maximal absorbance value). The substrate concentration in the first experiment was 1mM based
on this time point. Therefore, one can vary the substrate concentration below and above 1mM to
achieve a series of values still in linear range.
s(Mm) V (mM/min)
0.1 0.04
0.2 0.03
0.4 0.05
0.8 0.08
1 0.05
2 0.09
4 0.09
0 0.5 1 1.5 2 2.5 3 3.5 4 4.5
0
0.01
0.02
0.03
0.04
0.05
0.06
0.07
0.08
0.09
0.1
Graph of rate (v) Vs substrate (s)
S(mM)
V (mM/min)
Using the Beer-Lambert Law, (A= eCl), concentration of product generated at each
substrate concentration can be calculate. The millimolar extinction coefficient e for the product is
18 cm-1. Therefore, using the value in the equation will provide a result for concentration C that
is in mM (millimole per liter). The rate of reaction V for all substrate concentrations [S] used can
be calculated by dividing the concentration by the time allotted (1minutes).
Enzyme kinetics 10
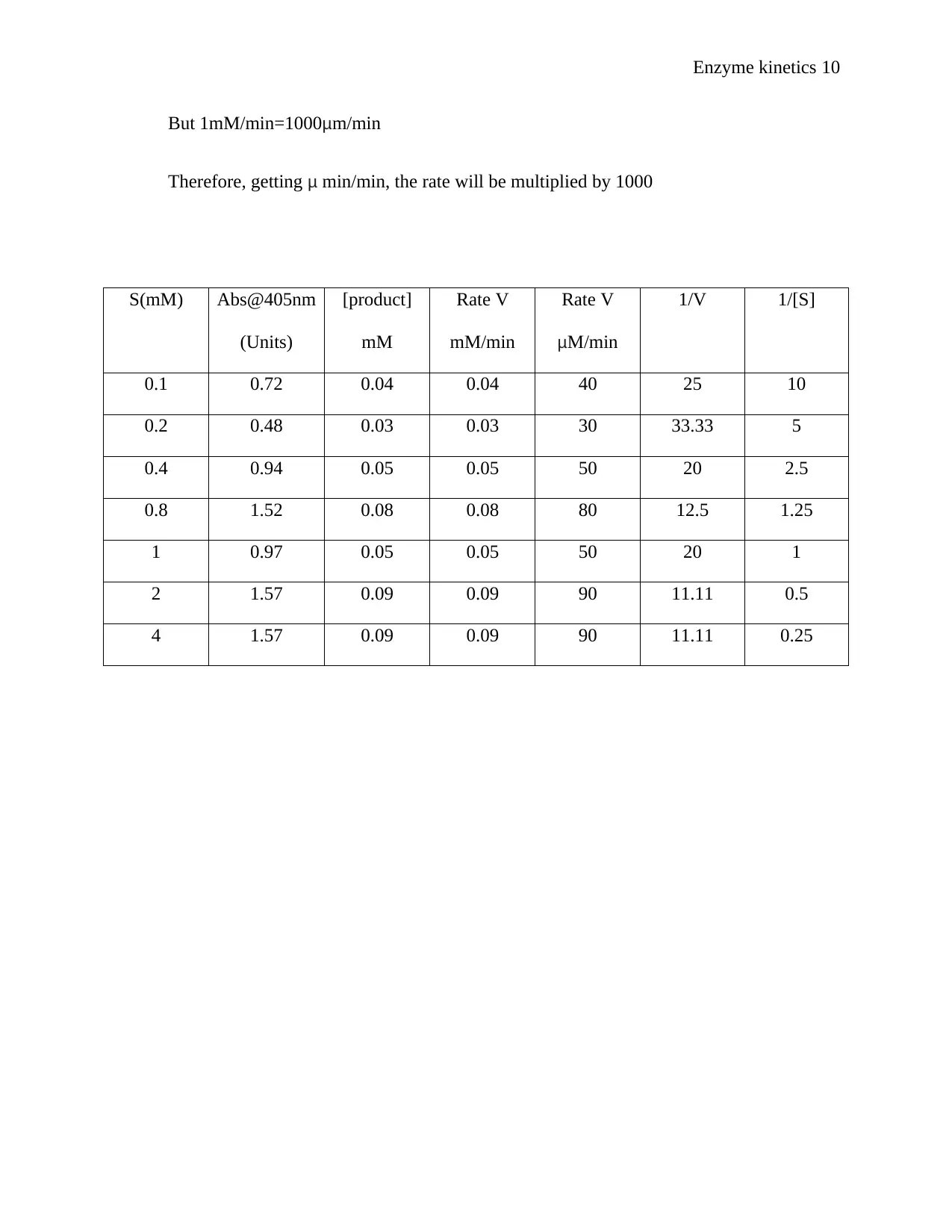
But 1mM/min=1000μm/min
Therefore, getting μ min/min, the rate will be multiplied by 1000
S(mM) Abs@405nm
(Units)
[product]
mM
Rate V
mM/min
Rate V
μM/min
1/V 1/[S]
0.1 0.72 0.04 0.04 40 25 10
0.2 0.48 0.03 0.03 30 33.33 5
0.4 0.94 0.05 0.05 50 20 2.5
0.8 1.52 0.08 0.08 80 12.5 1.25
1 0.97 0.05 0.05 50 20 1
2 1.57 0.09 0.09 90 11.11 0.5
4 1.57 0.09 0.09 90 11.11 0.25
But 1mM/min=1000μm/min
Therefore, getting μ min/min, the rate will be multiplied by 1000
S(mM) Abs@405nm
(Units)
[product]
mM
Rate V
mM/min
Rate V
μM/min
1/V 1/[S]
0.1 0.72 0.04 0.04 40 25 10
0.2 0.48 0.03 0.03 30 33.33 5
0.4 0.94 0.05 0.05 50 20 2.5
0.8 1.52 0.08 0.08 80 12.5 1.25
1 0.97 0.05 0.05 50 20 1
2 1.57 0.09 0.09 90 11.11 0.5
4 1.57 0.09 0.09 90 11.11 0.25
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.
Enzyme kinetics 11
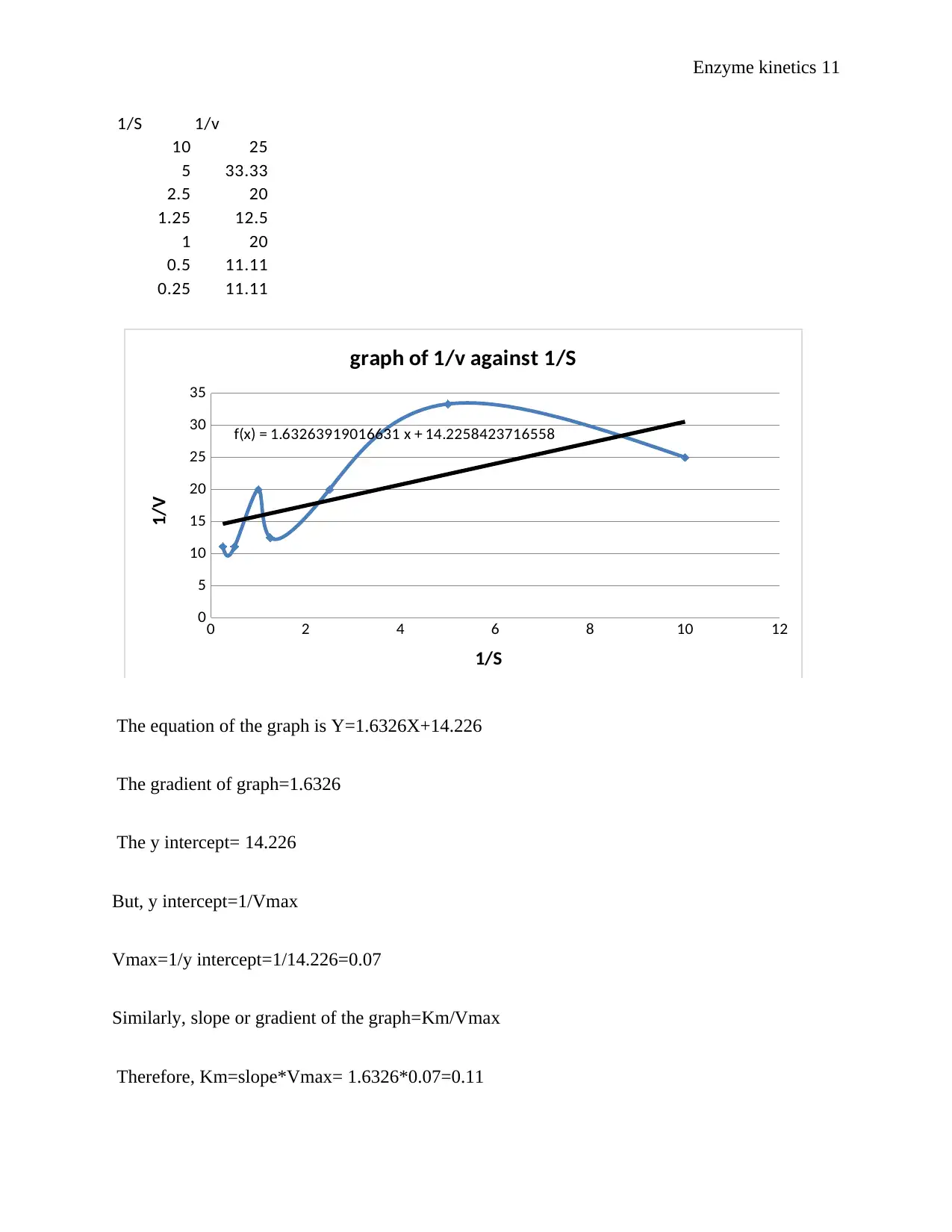
1/S 1/v
10 25
5 33.33
2.5 20
1.25 12.5
1 20
0.5 11.11
0.25 11.11
0 2 4 6 8 10 12
0
5
10
15
20
25
30
35
f(x) = 1.63263919016631 x + 14.2258423716558
graph of 1/v against 1/S
1/S
1/V
The equation of the graph is Y=1.6326X+14.226
The gradient of graph=1.6326
The y intercept= 14.226
But, y intercept=1/Vmax
Vmax=1/y intercept=1/14.226=0.07
Similarly, slope or gradient of the graph=Km/Vmax
Therefore, Km=slope*Vmax= 1.6326*0.07=0.11
1/S 1/v
10 25
5 33.33
2.5 20
1.25 12.5
1 20
0.5 11.11
0.25 11.11
0 2 4 6 8 10 12
0
5
10
15
20
25
30
35
f(x) = 1.63263919016631 x + 14.2258423716558
graph of 1/v against 1/S
1/S
1/V
The equation of the graph is Y=1.6326X+14.226
The gradient of graph=1.6326
The y intercept= 14.226
But, y intercept=1/Vmax
Vmax=1/y intercept=1/14.226=0.07
Similarly, slope or gradient of the graph=Km/Vmax
Therefore, Km=slope*Vmax= 1.6326*0.07=0.11

Enzyme kinetics 12
The Michaelis-Menten equation hold only when [s]>>> [E], when a steady-state concentration of
ES may be accomplished, and only for the first velocities. Under these conditions, the substrate
concentration may be denoted as [S] 0, the first substrate concentration. Moreover, at t=0, and
shortly afterwards, the p concentration will be trivial, and therefore the prospect that the reverse
reaction will interfere is eradicated. The Michaelis-Menten equation has change in numerous
ways to create the straight line equation. In the Line weaver-Burk plot, straight line is formed
with x intercept as -1/km, y intercept as 1/Vmax and slope=Km/Vmax. The above linear
transformation may be used in the determination of Km and Vmax for an enzyme, which shows
the enzyme affinity for its substrate and the catalytic efficacy of the enzyme.
The Michaelis-Menten equation hold only when [s]>>> [E], when a steady-state concentration of
ES may be accomplished, and only for the first velocities. Under these conditions, the substrate
concentration may be denoted as [S] 0, the first substrate concentration. Moreover, at t=0, and
shortly afterwards, the p concentration will be trivial, and therefore the prospect that the reverse
reaction will interfere is eradicated. The Michaelis-Menten equation has change in numerous
ways to create the straight line equation. In the Line weaver-Burk plot, straight line is formed
with x intercept as -1/km, y intercept as 1/Vmax and slope=Km/Vmax. The above linear
transformation may be used in the determination of Km and Vmax for an enzyme, which shows
the enzyme affinity for its substrate and the catalytic efficacy of the enzyme.
1 out of 12
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
© 2024 | Zucol Services PVT LTD | All rights reserved.
