GENETICS 13 Assignment: Molecular Biology and Genetic Processes
VerifiedAdded on 2022/10/15
|15
|2901
|21
Homework Assignment
AI Summary
This genetics assignment delves into various aspects of molecular biology and genetics. It begins with an overview of RNA polymerase II and retroviruses, followed by explanations of 5’Cap function, kinase enzymes, and structural differences between chlorophyll A and B. The assignment then covers topics such as cell ploidy, DNA and RNA binding, and the composition of organisms. It further explores lipid assembly in aqueous solutions, the differences between RNA and DNA structures, and the genetic concepts of incomplete and co-dominance, along with phenotype and genotype ratios. The document also examines the probability calculations in genetic crosses, the roles of promoters and start codons, and the process of DNA replication. It then proceeds to glycolysis, detailing its first and third steps, and explores the maladaptive nature of RuBisCo. The assignment also touches upon functional groups, evolutionary trees, and the biochemical mechanisms of RNA splicing, including the roles of snRNAs and snRNPs. Finally, it describes the three steps required to convert pre-mRNA to mature RNA in eukaryotes and discusses the longevity of eukaryotic mRNA compared to prokaryotic mRNA. The assignment concludes with a list of terms related to DNA replication and a brief overview of transcription initiation in bacteria.

Running head: GENETICS
Genetics
Name of Student:
Name of University:
Author’s Note:
Genetics
Name of Student:
Name of University:
Author’s Note:
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

1GENETICS
1. RNA polymerase II
2. An exception to central dogma of molecular biology is retrovirus which transcribes RNA
into DNA.
3. The core RNA polymerase does not contain sigma catalytic unit and holoenzyme carries
the sigma factor which is needed for initiation of transcription.
4. The main function of 5’Cap to protect the end of mRNA from methylation and
degradation from exonucleases.
5. Kinase referred to those enzymes which catalyse the transfer of phosphate group to other
enzymes for activation or deactivation.
6. The main structural difference between chlorophyll A and chlorophyll B is carrier of
functional group at 3rd carbon. In chlorophyll A 3rd carbon contain methyl group while
chlorophyll contain aldehyde group.
7. If a cell is treated with α-amanitin, the transcription factor and other proteins become
degraded and transcription process is halted.
8. When diploid strain 2x is being crossed with tetraploid strain 4x, in process of
fertilisation, haploid number of DNA from 2x which is 1x and haploid number of 4x
strain which is 2x are fused, thus give rise to 3x triploid organism.
9. Reason for occurrence of over 7000 nucleus in a cell is lack of cytokinesis during
mitosis.
10. If there is absence of non-coding regions, the DNA and RNA will not bind at non-coding
region and in micrograph large tangling loops will be observed.
11. Ninety six percent of every organism is made up of oxygen, carbon, hydrogen and
nitrogen.
1. RNA polymerase II
2. An exception to central dogma of molecular biology is retrovirus which transcribes RNA
into DNA.
3. The core RNA polymerase does not contain sigma catalytic unit and holoenzyme carries
the sigma factor which is needed for initiation of transcription.
4. The main function of 5’Cap to protect the end of mRNA from methylation and
degradation from exonucleases.
5. Kinase referred to those enzymes which catalyse the transfer of phosphate group to other
enzymes for activation or deactivation.
6. The main structural difference between chlorophyll A and chlorophyll B is carrier of
functional group at 3rd carbon. In chlorophyll A 3rd carbon contain methyl group while
chlorophyll contain aldehyde group.
7. If a cell is treated with α-amanitin, the transcription factor and other proteins become
degraded and transcription process is halted.
8. When diploid strain 2x is being crossed with tetraploid strain 4x, in process of
fertilisation, haploid number of DNA from 2x which is 1x and haploid number of 4x
strain which is 2x are fused, thus give rise to 3x triploid organism.
9. Reason for occurrence of over 7000 nucleus in a cell is lack of cytokinesis during
mitosis.
10. If there is absence of non-coding regions, the DNA and RNA will not bind at non-coding
region and in micrograph large tangling loops will be observed.
11. Ninety six percent of every organism is made up of oxygen, carbon, hydrogen and
nitrogen.

2GENETICS
12. Ile-Gly-Gly-Phe-Asp-Leu-Met-Pro-Arg-Leu.
13. Lipids can assemble in aqueous solution in the following two ways:
They can assemble into bilayers which forms sealed compartments and then these
compartments reseal when torn. The hydrophilic molecules dissolve readily in water
as they have charged groups, whereas hydrophobic molecules are insoluble in water
as all the atoms are uncharged.
Another way by which lipids assemble in water includes by reorganization of
adjacent water molecules into ice like cages that come and surround the hydrophobic
molecule.
14. The difference between the structure of RNA and DNA is depicted in the table below:
RNA DNA
It is single stranded structure and contain
ribose sugar.
It is double standard structure and contain
deoxyribose sugar at 2 end.
Adenine always pair with Uracil and
cytosine to guanine.
Adenine pairs up with thymine and
cytosine to guanine.
15. The fewest number of based that could code for 14 different amino acids includes 2
bases.
16. Incomplete dominance is a type of inheritance in which neither of the allele express
themselves independently. Hence, a third phenotype is expressed which is a blend of both
alleles. For example, when a pure red coloured flower is crossed with white coloured
flower, the resultant phenotype in the first generation includes pink flower
(heterozygous). In contrast, co-dominance is a type of inheritance in which one version of
12. Ile-Gly-Gly-Phe-Asp-Leu-Met-Pro-Arg-Leu.
13. Lipids can assemble in aqueous solution in the following two ways:
They can assemble into bilayers which forms sealed compartments and then these
compartments reseal when torn. The hydrophilic molecules dissolve readily in water
as they have charged groups, whereas hydrophobic molecules are insoluble in water
as all the atoms are uncharged.
Another way by which lipids assemble in water includes by reorganization of
adjacent water molecules into ice like cages that come and surround the hydrophobic
molecule.
14. The difference between the structure of RNA and DNA is depicted in the table below:
RNA DNA
It is single stranded structure and contain
ribose sugar.
It is double standard structure and contain
deoxyribose sugar at 2 end.
Adenine always pair with Uracil and
cytosine to guanine.
Adenine pairs up with thymine and
cytosine to guanine.
15. The fewest number of based that could code for 14 different amino acids includes 2
bases.
16. Incomplete dominance is a type of inheritance in which neither of the allele express
themselves independently. Hence, a third phenotype is expressed which is a blend of both
alleles. For example, when a pure red coloured flower is crossed with white coloured
flower, the resultant phenotype in the first generation includes pink flower
(heterozygous). In contrast, co-dominance is a type of inheritance in which one version of
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

3GENETICS
the allele from each parent is expressed. Hence, phenotypes of both alleles are expressed.
The F1 generation phenotypes have resemblance with both its parents.
17. The following table depicts the phenotype and genotype ratios between Yy and yy pea
individuals:
Y- Dominant yellow seed colour and y – Recessive green colour
The phenotypic ration is 2 (yellow seed): 2 (green seed)
The genotypic ratio is 2 (Yy) :2 (yy)
18.
In AABbCc × AaBbCc cross
The probability of having AaBbCc is 1/2 × 1/2 × 1/2 = 1/8
As in AA × Aa 1/2 is the probability of having Aa genotype
In Bb× Bb 1/2 is the probability of having Bb genotype.
In Cc × Cc 1/2 is the probability of having Cc genotype.
19. In cross of AaBbCc × AaBbCc
Y y
y Yy (yellow seed) yy (green seed)
y Yy (yellow seed) Yy (green seed)
the allele from each parent is expressed. Hence, phenotypes of both alleles are expressed.
The F1 generation phenotypes have resemblance with both its parents.
17. The following table depicts the phenotype and genotype ratios between Yy and yy pea
individuals:
Y- Dominant yellow seed colour and y – Recessive green colour
The phenotypic ration is 2 (yellow seed): 2 (green seed)
The genotypic ratio is 2 (Yy) :2 (yy)
18.
In AABbCc × AaBbCc cross
The probability of having AaBbCc is 1/2 × 1/2 × 1/2 = 1/8
As in AA × Aa 1/2 is the probability of having Aa genotype
In Bb× Bb 1/2 is the probability of having Bb genotype.
In Cc × Cc 1/2 is the probability of having Cc genotype.
19. In cross of AaBbCc × AaBbCc
Y y
y Yy (yellow seed) yy (green seed)
y Yy (yellow seed) Yy (green seed)
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

4GENETICS
The probability of having homozygous recessive trait of genotype aabbcc will be
1/4 × 1/4 × 1/4= 1/64
20. Promoters are the regions of DNA from where transcription starts, whereas start codons
are the first base that is translated in mRNA. The start codon and promote are similar to
each other. Hence, the similarity between the two is that both acts as successful initiators.
The promoter sequence is necessary for successful initiation of transcription, whereas the
start codon is necessary to successfully initiate translation. The difference between the
two terms is that promoters executes its function on an mRNA, whereas starts codons are
parts of a DNA. The promoter helps in identifying the site from where translation can
take place, whereas starts codons is itself the region from where transcription process
starts.
21. In the process of DNA replication, it starts to synthesize at the 5’end of the DNA which
requires helicase enzyme to unbind the strand of DNA. SSB proteins binds at each of the
strand to keep them separated. As the result of it, one strand orient in 3’ to 5’ directing
known as leading strands and other orient at 5’to 3’ known as lagging strands. Primer are
then attached at the 5’ end of each strand with help to DNA polymerase new DNA are
synthesise. At lagging strand though it orient in opposite direction, various primer binds
at fragment of lagging strand and synthesis takes place at 5’ to 3’ direction. The
fragments of DNA are joined by ligase enzyme and form full strand of DNA by clamp
loading mechanism.
22. The first step of glycolysis involves breakdown of glucose to glucose-6-phosphate by
phosphorylation of ATP molecule by Hexokinase enzyme. The step is significant because
glucose cannot be further metabolized until it is broken to glucose-6-phosphate. This step
The probability of having homozygous recessive trait of genotype aabbcc will be
1/4 × 1/4 × 1/4= 1/64
20. Promoters are the regions of DNA from where transcription starts, whereas start codons
are the first base that is translated in mRNA. The start codon and promote are similar to
each other. Hence, the similarity between the two is that both acts as successful initiators.
The promoter sequence is necessary for successful initiation of transcription, whereas the
start codon is necessary to successfully initiate translation. The difference between the
two terms is that promoters executes its function on an mRNA, whereas starts codons are
parts of a DNA. The promoter helps in identifying the site from where translation can
take place, whereas starts codons is itself the region from where transcription process
starts.
21. In the process of DNA replication, it starts to synthesize at the 5’end of the DNA which
requires helicase enzyme to unbind the strand of DNA. SSB proteins binds at each of the
strand to keep them separated. As the result of it, one strand orient in 3’ to 5’ directing
known as leading strands and other orient at 5’to 3’ known as lagging strands. Primer are
then attached at the 5’ end of each strand with help to DNA polymerase new DNA are
synthesise. At lagging strand though it orient in opposite direction, various primer binds
at fragment of lagging strand and synthesis takes place at 5’ to 3’ direction. The
fragments of DNA are joined by ligase enzyme and form full strand of DNA by clamp
loading mechanism.
22. The first step of glycolysis involves breakdown of glucose to glucose-6-phosphate by
phosphorylation of ATP molecule by Hexokinase enzyme. The step is significant because
glucose cannot be further metabolized until it is broken to glucose-6-phosphate. This step
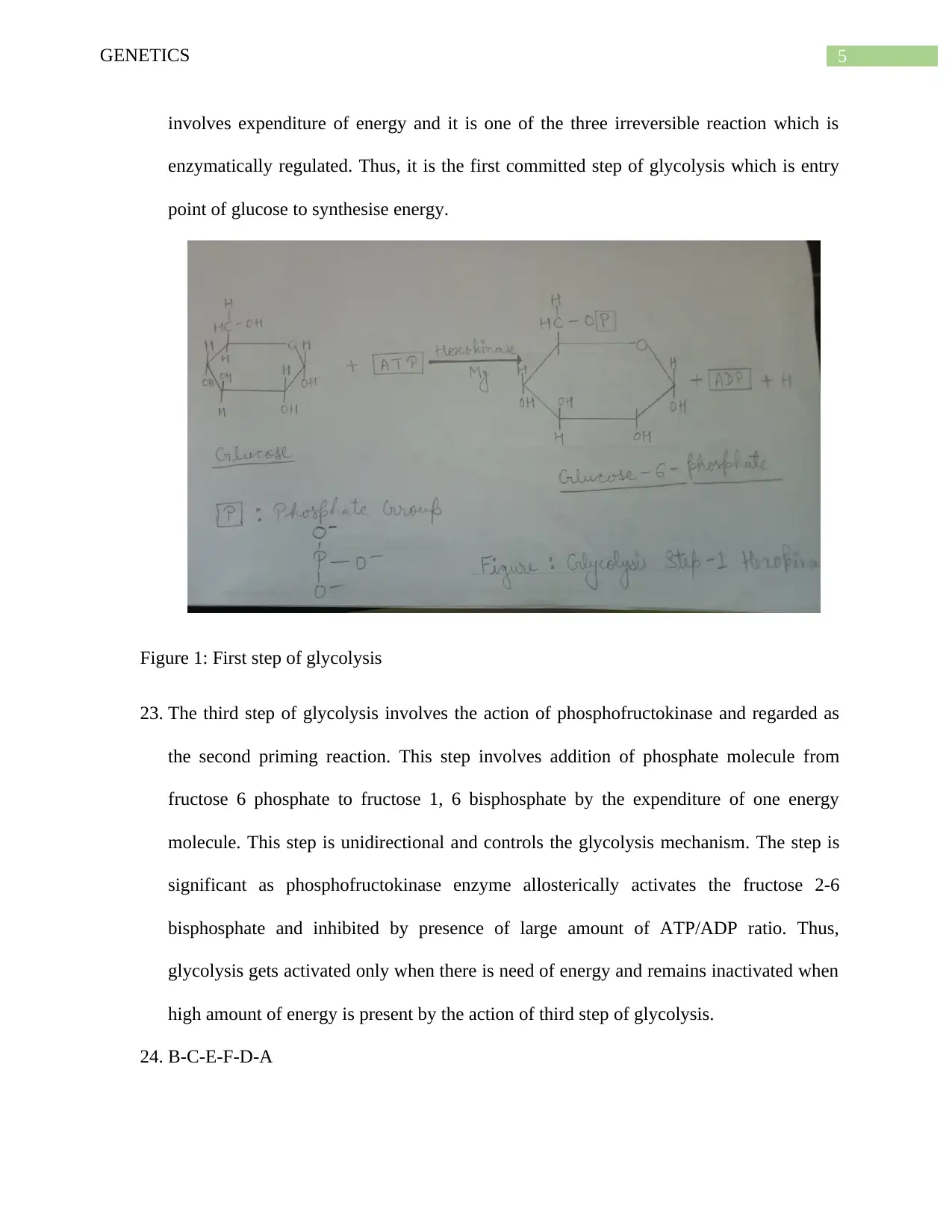
5GENETICS
involves expenditure of energy and it is one of the three irreversible reaction which is
enzymatically regulated. Thus, it is the first committed step of glycolysis which is entry
point of glucose to synthesise energy.
Figure 1: First step of glycolysis
23. The third step of glycolysis involves the action of phosphofructokinase and regarded as
the second priming reaction. This step involves addition of phosphate molecule from
fructose 6 phosphate to fructose 1, 6 bisphosphate by the expenditure of one energy
molecule. This step is unidirectional and controls the glycolysis mechanism. The step is
significant as phosphofructokinase enzyme allosterically activates the fructose 2-6
bisphosphate and inhibited by presence of large amount of ATP/ADP ratio. Thus,
glycolysis gets activated only when there is need of energy and remains inactivated when
high amount of energy is present by the action of third step of glycolysis.
24. B-C-E-F-D-A
involves expenditure of energy and it is one of the three irreversible reaction which is
enzymatically regulated. Thus, it is the first committed step of glycolysis which is entry
point of glucose to synthesise energy.
Figure 1: First step of glycolysis
23. The third step of glycolysis involves the action of phosphofructokinase and regarded as
the second priming reaction. This step involves addition of phosphate molecule from
fructose 6 phosphate to fructose 1, 6 bisphosphate by the expenditure of one energy
molecule. This step is unidirectional and controls the glycolysis mechanism. The step is
significant as phosphofructokinase enzyme allosterically activates the fructose 2-6
bisphosphate and inhibited by presence of large amount of ATP/ADP ratio. Thus,
glycolysis gets activated only when there is need of energy and remains inactivated when
high amount of energy is present by the action of third step of glycolysis.
24. B-C-E-F-D-A
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

6GENETICS
25. RuBisCo is maladaptive enzyme because it functions both as oxygenase and
carboxygenase which is influenced by the atmosphere. The function of RuBisCo can be
gathered from photorespiration. Three reason for which it is regarded as maladaptive
enzyme are:
When there is low concentration of CO2 inside the leaf or high concentration of
oxygen, the enzyme functions as oxygenase and grabs oxygen instead of carbon
dioxide.
When there is high concentration of carbon dioxide it stats to function as
carboxygenase and grab CO2 resulting in formation of energy.
RuBisCo does not function well in very high and very low temperature.
26. Functional groups are groups of atoms that are seen in organic molecules and that provide
specific chemical properties to the organic molecules. Such groups are mostly found
along the carbon backbone. Different types of macromolecules include proteins, lipids,
nucleic acids and carbohydrates and each one of them have a functional group that
contribute to different chemical properties of the macromolecules. The four major types
of functional groups are as follows:
1. Hydroxyl: This groups has polar properties and simple structure characterised by
presence of H and O (R OH)
2. Methyl: It is a non-polar molecule with presence of C and H (R CH3)
3. Carbonyl: It is polar molecule with double bond to oxygen and characterised by central C
and O
4. Phosphate: It is acidic in nature and has a complex structure characterised by presence of
P.
25. RuBisCo is maladaptive enzyme because it functions both as oxygenase and
carboxygenase which is influenced by the atmosphere. The function of RuBisCo can be
gathered from photorespiration. Three reason for which it is regarded as maladaptive
enzyme are:
When there is low concentration of CO2 inside the leaf or high concentration of
oxygen, the enzyme functions as oxygenase and grabs oxygen instead of carbon
dioxide.
When there is high concentration of carbon dioxide it stats to function as
carboxygenase and grab CO2 resulting in formation of energy.
RuBisCo does not function well in very high and very low temperature.
26. Functional groups are groups of atoms that are seen in organic molecules and that provide
specific chemical properties to the organic molecules. Such groups are mostly found
along the carbon backbone. Different types of macromolecules include proteins, lipids,
nucleic acids and carbohydrates and each one of them have a functional group that
contribute to different chemical properties of the macromolecules. The four major types
of functional groups are as follows:
1. Hydroxyl: This groups has polar properties and simple structure characterised by
presence of H and O (R OH)
2. Methyl: It is a non-polar molecule with presence of C and H (R CH3)
3. Carbonyl: It is polar molecule with double bond to oxygen and characterised by central C
and O
4. Phosphate: It is acidic in nature and has a complex structure characterised by presence of
P.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

7GENETICS
27. Evolutionary tree
Figure 2: Evolutional tree for Bacteria, archaea and Eukarya
28. The biochemical mechanism of RNA splicing occurs with the objective to remove intron
which is the non-coding region of DNA. The mechanism has made use of many enzyme
and protein to cleave intron from the heterogeneous RNA. The initiation of RNA splicing
start at the splice site which is located at the 5 and 3 ends of introns. The removal of
intron begins at its 5’ end which contains GU dinucleotide and it ends at the AG at 3’
end. The sequences are consensus and know to be significant because any change in
sequence will result in inhibition of the process.
The main components of splicing apparatus in the intron are known as snRNA which
consists of U1, U2. U4, U5 and U6. These RNA molecules in association with the protein are
known as snRNPs. At the first step of RNA splicing, U1-snRNP bind at the 5’ splice side, branch
point binding protein binds at the branch point and U2AF interact with the polypyrimidine tract.
At this step first transesterification reaction occur where the 5 end of the intron are cleaved and it
is promoted by the OH group which is attached at the 2 carbon of Adenine situated in intron
27. Evolutionary tree
Figure 2: Evolutional tree for Bacteria, archaea and Eukarya
28. The biochemical mechanism of RNA splicing occurs with the objective to remove intron
which is the non-coding region of DNA. The mechanism has made use of many enzyme
and protein to cleave intron from the heterogeneous RNA. The initiation of RNA splicing
start at the splice site which is located at the 5 and 3 ends of introns. The removal of
intron begins at its 5’ end which contains GU dinucleotide and it ends at the AG at 3’
end. The sequences are consensus and know to be significant because any change in
sequence will result in inhibition of the process.
The main components of splicing apparatus in the intron are known as snRNA which
consists of U1, U2. U4, U5 and U6. These RNA molecules in association with the protein are
known as snRNPs. At the first step of RNA splicing, U1-snRNP bind at the 5’ splice side, branch
point binding protein binds at the branch point and U2AF interact with the polypyrimidine tract.
At this step first transesterification reaction occur where the 5 end of the intron are cleaved and it
is promoted by the OH group which is attached at the 2 carbon of Adenine situated in intron

8GENETICS
sequence. The OH group attacks and results in formation of 5’-2’ phosphodiester bond by
linking the first nucleotide of the intron with the internal adenosine. Thus mechanism results in
formation of looped structure which is known as lariat structure. In the second step, there is
addition of U2 snRNP at the branch point and it bring the 5’ splice site in close proximity of the
branch point. Next, the contribution of U4/U6 at the branch point form the spliceosome. Next
participation of the U5 protein at the 3’end of the intron bring it in close proximity of the 5’end
and intron is cut and joined by the second transesterification reaction. In this, OH group at the 3
end of upstream exon attacks the phosphodiester bond which is present at the 3 splice site.
Hence, it removes the intron in the form of lariat structure. The exon released are joins the newly
formed 5 end of the downstream exon and complete the splicing process.
Figure 3: A diagram depicting how introns are removed by RNA splicing
29. In eukaryotes the three steps which is required to convert pre-MRNA to mature RNA are
the following:
sequence. The OH group attacks and results in formation of 5’-2’ phosphodiester bond by
linking the first nucleotide of the intron with the internal adenosine. Thus mechanism results in
formation of looped structure which is known as lariat structure. In the second step, there is
addition of U2 snRNP at the branch point and it bring the 5’ splice site in close proximity of the
branch point. Next, the contribution of U4/U6 at the branch point form the spliceosome. Next
participation of the U5 protein at the 3’end of the intron bring it in close proximity of the 5’end
and intron is cut and joined by the second transesterification reaction. In this, OH group at the 3
end of upstream exon attacks the phosphodiester bond which is present at the 3 splice site.
Hence, it removes the intron in the form of lariat structure. The exon released are joins the newly
formed 5 end of the downstream exon and complete the splicing process.
Figure 3: A diagram depicting how introns are removed by RNA splicing
29. In eukaryotes the three steps which is required to convert pre-MRNA to mature RNA are
the following:
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

9GENETICS
5’ capping – at this step, the pre-RNA are capped at the 5 end which consist of 7
methyl-guanosine residue and make 5-5 triphosphate bond. The enzymes involved
in the reaction are phosphohydralase, guanyly transferase and guanine 7 methyl
transferase. The whole reaction is synthesized by the RNA polymerase II.
3 tailing – This step is also known as polyadenylation where at the 3 end of RNA
10-200 adenine molecule is added by the action of poly (A) polymerase. The main
function of polyadenylation is to export the mature MRNA from the nucleus and
maintain stability of the MRNA.
Splicing of RNA- the last step of the mechanism, is splicing of RNA molecule
where the introns are removed from the pre-MRNA and exon are joined by
covalent bond. The resulting sequence which constitute of exon is known as
mature RNA.
Eukaryotic MRNA would last longer than the prokaryotic MRNA because in
prokaryotes, the cell lacks nucleus and the mechanism of transcription and translation occur at
simultaneously in the cell. As the result of this, the MRNA is having very short half live of 3
minutes and gets degraded by the action of exonuclease which starts at the 3 end of the RNA
molecule. Like prokaryotes, eukaryotic MRNA also gets degraded following the mechanism of
protein synthesis but takes longer time than the prokaryotic RNA because they have longer half-
live which is around 30 minutes.
30. The answers are as follow:
a) Origin of replication
b) Leading strand
c) Lagging strand
5’ capping – at this step, the pre-RNA are capped at the 5 end which consist of 7
methyl-guanosine residue and make 5-5 triphosphate bond. The enzymes involved
in the reaction are phosphohydralase, guanyly transferase and guanine 7 methyl
transferase. The whole reaction is synthesized by the RNA polymerase II.
3 tailing – This step is also known as polyadenylation where at the 3 end of RNA
10-200 adenine molecule is added by the action of poly (A) polymerase. The main
function of polyadenylation is to export the mature MRNA from the nucleus and
maintain stability of the MRNA.
Splicing of RNA- the last step of the mechanism, is splicing of RNA molecule
where the introns are removed from the pre-MRNA and exon are joined by
covalent bond. The resulting sequence which constitute of exon is known as
mature RNA.
Eukaryotic MRNA would last longer than the prokaryotic MRNA because in
prokaryotes, the cell lacks nucleus and the mechanism of transcription and translation occur at
simultaneously in the cell. As the result of this, the MRNA is having very short half live of 3
minutes and gets degraded by the action of exonuclease which starts at the 3 end of the RNA
molecule. Like prokaryotes, eukaryotic MRNA also gets degraded following the mechanism of
protein synthesis but takes longer time than the prokaryotic RNA because they have longer half-
live which is around 30 minutes.
30. The answers are as follow:
a) Origin of replication
b) Leading strand
c) Lagging strand
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

10GENETICS
d) Okazaki
e) Primase (dnaG)
f) DNA polymerase I
g) Exonuclease
h) Ligase
31. Transcription initiation in bacteria consists of the three steps of initiation, elongation and
termination: Initiation: The process of transcription initiates with the action of the RNA polymerase
enzyme that binds to the promoter region. The promoter has DNA sequences that help the
RNA polymerase to bind with the enzymes and form the transcription bubble. The
binding of the RNA polymerase helps to form a closed complex. In addition, an open
complex is formed when the RNA polymerase enzymes separates 10-14 bases. This is
followed by synthesis of nucleotides. Elongation: The next step of elongation initiates when the RNA polymerase recognizes
the promoter. In this phase, the elongation of the RNA strand occurs by the addition of
the new nucleotides. The process of elongation takes places in the 3' to 5' direction. The
resulting transcript is similar to the non-template strand. However, each T is replaced
with a U in the RNA transcript.
Termination: The process of termination takes place in bacteria by means of rho-
dependent termination and rho-independent termination. Rho dependent termination
takes place when the RNA contains a rho factors that binds with the single strander RNA
that exit from the polymerase enzyme. After meeting the polymerase enzyme at the
transcription bubble, the Rho pulls the RNA transcript and template DNA standard apart
d) Okazaki
e) Primase (dnaG)
f) DNA polymerase I
g) Exonuclease
h) Ligase
31. Transcription initiation in bacteria consists of the three steps of initiation, elongation and
termination: Initiation: The process of transcription initiates with the action of the RNA polymerase
enzyme that binds to the promoter region. The promoter has DNA sequences that help the
RNA polymerase to bind with the enzymes and form the transcription bubble. The
binding of the RNA polymerase helps to form a closed complex. In addition, an open
complex is formed when the RNA polymerase enzymes separates 10-14 bases. This is
followed by synthesis of nucleotides. Elongation: The next step of elongation initiates when the RNA polymerase recognizes
the promoter. In this phase, the elongation of the RNA strand occurs by the addition of
the new nucleotides. The process of elongation takes places in the 3' to 5' direction. The
resulting transcript is similar to the non-template strand. However, each T is replaced
with a U in the RNA transcript.
Termination: The process of termination takes place in bacteria by means of rho-
dependent termination and rho-independent termination. Rho dependent termination
takes place when the RNA contains a rho factors that binds with the single strander RNA
that exit from the polymerase enzyme. After meeting the polymerase enzyme at the
transcription bubble, the Rho pulls the RNA transcript and template DNA standard apart

11GENETICS
thus ending the transcription process. In contrast, rho-independent termination takes place
when RNA polymerase approaches a region rich in C and G nucleotides. The RNA that is
transcribed from this region folds back on itself and result in formation of a hair pin like
structure. This process halts the actions of polymerase and results in termination of the
transcription process.
32. The tertiary structure of a protein consists of a single polypeptide structure followed by
one or more secondary structures. The five important types of R-group interactions in
tertiary proteins includes the following:
Hydrogen bonding: In this case, polar R groups orient with other polar R groups
Ionic bonding: In this case, all positively charged R groups bind with each other
Hydrophobic interactions: Amino acids orients towards centre of the polypeptide
resulting in avoidance of water.
Hydrophilic interactions: In this interaction, amino acid orient outwards to remain close
to water
Disulphide bridges: In this case, R group forms a bond with another cysteine
33.
thus ending the transcription process. In contrast, rho-independent termination takes place
when RNA polymerase approaches a region rich in C and G nucleotides. The RNA that is
transcribed from this region folds back on itself and result in formation of a hair pin like
structure. This process halts the actions of polymerase and results in termination of the
transcription process.
32. The tertiary structure of a protein consists of a single polypeptide structure followed by
one or more secondary structures. The five important types of R-group interactions in
tertiary proteins includes the following:
Hydrogen bonding: In this case, polar R groups orient with other polar R groups
Ionic bonding: In this case, all positively charged R groups bind with each other
Hydrophobic interactions: Amino acids orients towards centre of the polypeptide
resulting in avoidance of water.
Hydrophilic interactions: In this interaction, amino acid orient outwards to remain close
to water
Disulphide bridges: In this case, R group forms a bond with another cysteine
33.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 15
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.

