Role of Microarray Comparative Genomic Hybridization
VerifiedAdded on 2023/01/13
|13
|3125
|69
AI Summary
The Microarray Comparative Genomic Hybridization (array CGH) is a modern molecular cytogenic technique useful for rapid analysis and evaluation of the whole genome with megabase resolution properties. The genetic technologies give an opportunity to achieve efficiency towards the diagnosis of several diseases as well as understanding their molecular basis. The technique helps in the study of thousands and millions of genomic loci at once and thus enabling accuracy in detecting the variations of the DNA copying.
Contribute Materials
Your contribution can guide someone’s learning journey. Share your
documents today.

Running head: GENOMIC HEALTH 1
Role of Microarray Comparative Genomic Hybridization
Student’s Name
Professor’s Name
Institution Affiliation
Date
Role of Microarray Comparative Genomic Hybridization
Student’s Name
Professor’s Name
Institution Affiliation
Date
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

GENOMIC HEALTH 2
Introduction
The Microarray Comparative Genomic Hybridization (array CGH) is a modern molecular
cytogenic technique useful for rapid analysis and evaluation of the whole genome with mega-
base resolution properties. The genetic technologies give an opportunity to achieve efficiency
towards the diagnosis of several diseases as well as understanding their molecular basis. The
technique helps in the study of thousands and millions of genomic loci at once and thus enabling
accuracy in detecting the variations of the DNA copying. The new microarray CGH technology
has been adopted in recent years and the most appropriate technique in clinical genetics. The
genomic disorders that result from the rearrangement of the DNA which involves a repeat of
region-specific DNA sequence due to the abnormal rearrangement of genomic fragments. The
array CGH technique has been the fastest growing technique due to its ability to detect thousands
and millions mosaic and non-mosaic aneuploidies, known micro-duplication syndromes,
unbalanced chromosomal rearrangements a subtelomeric imbalance. This new technique has
improved the clinical genetic examination from 10 – 16% in handling cases of patients with the
genomic disorder to normal karyotype. The microarray CGH has high power to uncover several
copy number variations (CNVs) of unknown clinical significance that is spread throughout the
human genome (Durmaz, Karaca, Demkow, Toruner, Schoumans & Cogulu, 2015).
Major advantages of array CGH over other genomic techniques
The emergence of the microarray cooperative genomic hybridization expressed great
significance and advantages to both physicians and the patients (Sadek & Mohamed, 2018).
First, the microarray CGH with multiplex formats enhances the simultaneous screening of
thousands of fully-characteristics of disease loci and their subtelomeric regions which shortened
Introduction
The Microarray Comparative Genomic Hybridization (array CGH) is a modern molecular
cytogenic technique useful for rapid analysis and evaluation of the whole genome with mega-
base resolution properties. The genetic technologies give an opportunity to achieve efficiency
towards the diagnosis of several diseases as well as understanding their molecular basis. The
technique helps in the study of thousands and millions of genomic loci at once and thus enabling
accuracy in detecting the variations of the DNA copying. The new microarray CGH technology
has been adopted in recent years and the most appropriate technique in clinical genetics. The
genomic disorders that result from the rearrangement of the DNA which involves a repeat of
region-specific DNA sequence due to the abnormal rearrangement of genomic fragments. The
array CGH technique has been the fastest growing technique due to its ability to detect thousands
and millions mosaic and non-mosaic aneuploidies, known micro-duplication syndromes,
unbalanced chromosomal rearrangements a subtelomeric imbalance. This new technique has
improved the clinical genetic examination from 10 – 16% in handling cases of patients with the
genomic disorder to normal karyotype. The microarray CGH has high power to uncover several
copy number variations (CNVs) of unknown clinical significance that is spread throughout the
human genome (Durmaz, Karaca, Demkow, Toruner, Schoumans & Cogulu, 2015).
Major advantages of array CGH over other genomic techniques
The emergence of the microarray cooperative genomic hybridization expressed great
significance and advantages to both physicians and the patients (Sadek & Mohamed, 2018).
First, the microarray CGH with multiplex formats enhances the simultaneous screening of
thousands of fully-characteristics of disease loci and their subtelomeric regions which shortened

GENOMIC HEALTH 3
the diagnostic process and minimized the cost as compared to the ordinary sequential
examinations that dealt with an individual locus at a time (Xavier et al, 2018). Second, the array
CGH is a modern discovery approach that can identify unknown clinical CNVs. It located the
genes making them the potential candidates who contributed to further studies resulting in the
discovery of genetic diseases. The technique also contributed much to the characterization of
genetic disorders. Thirdly, the array CGH technical development based on bacterial artificial
chromosomes (BAC) towards the oligonucleotide discovery. The more improved microarray
devices resolution enhanced significance in better understanding and definition of the genomic
losses and achievements. The appropriate definition of the correct genetic information within the
deleted/duplicated locus is of importance for efficient clinical management for featuring the
involved genes in the patient. Forth, the array CGH offers a wide rapid-genome analysis at very
high resolutions, and it provides contents that is directly connected to the genetic and physical
maps of the human genome. Fifth, the technique has been successfully used in the analysis of
cell lines and tumor samples and for high resolution in constitutional abnormalities examination
comprising of the single copy losses and gains in particular chromosomal locus or telomeres
(Rosenthal, Bernhisel, Brown, Kidd & Manley, 2017).
In a study using the array CGH in the wide genome to investigate patients with learning
abnormalities and dysmorphic, the following result was detected. Five copy number
alterations--- whereby one involved the telomere rearrangement and two were de novo which
were in the 20 patients resulting in a diagnostic potential of 15% (Giménez et al, 2015). In a
further study to investigate the presence of subtle DNA copy number variance using the
microarray CGH in 50 children with dysmorphism and learning disabilities, the DNA microarray
made from numerous clones that were spaced at an interval of 1 mb across the genome. There
the diagnostic process and minimized the cost as compared to the ordinary sequential
examinations that dealt with an individual locus at a time (Xavier et al, 2018). Second, the array
CGH is a modern discovery approach that can identify unknown clinical CNVs. It located the
genes making them the potential candidates who contributed to further studies resulting in the
discovery of genetic diseases. The technique also contributed much to the characterization of
genetic disorders. Thirdly, the array CGH technical development based on bacterial artificial
chromosomes (BAC) towards the oligonucleotide discovery. The more improved microarray
devices resolution enhanced significance in better understanding and definition of the genomic
losses and achievements. The appropriate definition of the correct genetic information within the
deleted/duplicated locus is of importance for efficient clinical management for featuring the
involved genes in the patient. Forth, the array CGH offers a wide rapid-genome analysis at very
high resolutions, and it provides contents that is directly connected to the genetic and physical
maps of the human genome. Fifth, the technique has been successfully used in the analysis of
cell lines and tumor samples and for high resolution in constitutional abnormalities examination
comprising of the single copy losses and gains in particular chromosomal locus or telomeres
(Rosenthal, Bernhisel, Brown, Kidd & Manley, 2017).
In a study using the array CGH in the wide genome to investigate patients with learning
abnormalities and dysmorphic, the following result was detected. Five copy number
alterations--- whereby one involved the telomere rearrangement and two were de novo which
were in the 20 patients resulting in a diagnostic potential of 15% (Giménez et al, 2015). In a
further study to investigate the presence of subtle DNA copy number variance using the
microarray CGH in 50 children with dysmorphism and learning disabilities, the DNA microarray
made from numerous clones that were spaced at an interval of 1 mb across the genome. There

GENOMIC HEALTH 4
were 12 patients which approximated to 12% of the total who represented 12 abnormalities of
copy number: 5 duplications with four copies inherited phenotypically and one de novo and
seven deletions composed of one inherited phenotypically and six de novo. The genes segments
that were altered ranged in size from single clones to large regions of 14 Mb. No recurrent
duplication or deletion resulted. According to the outcomes, it was concluded that the array CGH
would be continually used in wide screening of genome to tests the rearrangement imbalance in
children with learning disabilities (Kurtovic-Kozaric et al, 2016).
The learning disability, congenital anomalies and growth abnormality disorders is
characterized by the imbalance of the constitutional chromosomes. The recurrent chromosomal
imbalance can be demonstrated by features like trisomies and can be investigated through
cytogenetic technique. The use of the cytogenic technique is not much efficient due to its limits
resolution properties and its unreliability to subtle number variance. The modern technique of
microarray comparative genomic hybridization (CGH) uses the metaphase stage of the
chromosome to solve such chromosomal imbalance. Its high sensitivity allows the detection of
the chromosomal deletion at a small interval as 3 Mb. In a certain study, patients with learning
disabilities and dysmorphic features, ten percent of the patients detected small
duplications/deletions through the CGH method although it was not yet acknowledged on the G-
hooped karyotype (Fragouli et al, 2015).
Genotype Data analysis and discussion
The figures and the microarray pictures below are representations of different studies
that demonstrated the efficiency and accuracy of the array CGH.
were 12 patients which approximated to 12% of the total who represented 12 abnormalities of
copy number: 5 duplications with four copies inherited phenotypically and one de novo and
seven deletions composed of one inherited phenotypically and six de novo. The genes segments
that were altered ranged in size from single clones to large regions of 14 Mb. No recurrent
duplication or deletion resulted. According to the outcomes, it was concluded that the array CGH
would be continually used in wide screening of genome to tests the rearrangement imbalance in
children with learning disabilities (Kurtovic-Kozaric et al, 2016).
The learning disability, congenital anomalies and growth abnormality disorders is
characterized by the imbalance of the constitutional chromosomes. The recurrent chromosomal
imbalance can be demonstrated by features like trisomies and can be investigated through
cytogenetic technique. The use of the cytogenic technique is not much efficient due to its limits
resolution properties and its unreliability to subtle number variance. The modern technique of
microarray comparative genomic hybridization (CGH) uses the metaphase stage of the
chromosome to solve such chromosomal imbalance. Its high sensitivity allows the detection of
the chromosomal deletion at a small interval as 3 Mb. In a certain study, patients with learning
disabilities and dysmorphic features, ten percent of the patients detected small
duplications/deletions through the CGH method although it was not yet acknowledged on the G-
hooped karyotype (Fragouli et al, 2015).
Genotype Data analysis and discussion
The figures and the microarray pictures below are representations of different studies
that demonstrated the efficiency and accuracy of the array CGH.
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.
GENOMIC HEALTH 5
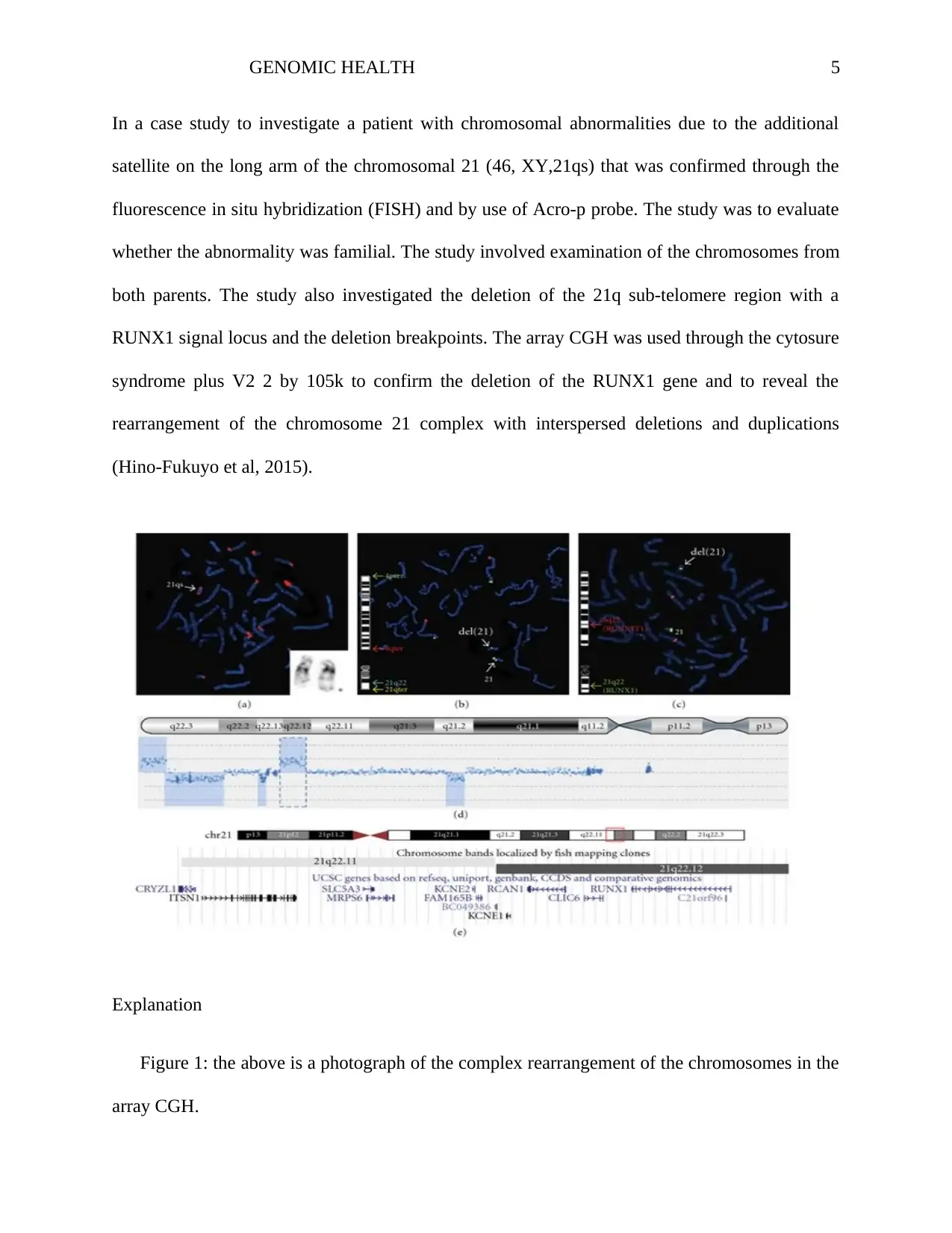
In a case study to investigate a patient with chromosomal abnormalities due to the additional
satellite on the long arm of the chromosomal 21 (46, XY,21qs) that was confirmed through the
fluorescence in situ hybridization (FISH) and by use of Acro-p probe. The study was to evaluate
whether the abnormality was familial. The study involved examination of the chromosomes from
both parents. The study also investigated the deletion of the 21q sub-telomere region with a
RUNX1 signal locus and the deletion breakpoints. The array CGH was used through the cytosure
syndrome plus V2 2 by 105k to confirm the deletion of the RUNX1 gene and to reveal the
rearrangement of the chromosome 21 complex with interspersed deletions and duplications
(Hino-Fukuyo et al, 2015).
Explanation
Figure 1: the above is a photograph of the complex rearrangement of the chromosomes in the
array CGH.
In a case study to investigate a patient with chromosomal abnormalities due to the additional
satellite on the long arm of the chromosomal 21 (46, XY,21qs) that was confirmed through the
fluorescence in situ hybridization (FISH) and by use of Acro-p probe. The study was to evaluate
whether the abnormality was familial. The study involved examination of the chromosomes from
both parents. The study also investigated the deletion of the 21q sub-telomere region with a
RUNX1 signal locus and the deletion breakpoints. The array CGH was used through the cytosure
syndrome plus V2 2 by 105k to confirm the deletion of the RUNX1 gene and to reveal the
rearrangement of the chromosome 21 complex with interspersed deletions and duplications
(Hino-Fukuyo et al, 2015).
Explanation
Figure 1: the above is a photograph of the complex rearrangement of the chromosomes in the
array CGH.

GENOMIC HEALTH 6
(a) The fluorescence in situ hybridization (FISH) study involved and acro-p probe that
targeted short arms of the acrocentric chromosomes to investigate the additional satellite
on the long arm with 21qs. The (symbol *) showed that the G-banded 21qs chromosome
got an additional satellite on its long arm.
(b) The study showed that the 21q22 chromosome region composed of RUNXI gene and the
21qter locus aligned the deleted chromosomes. The deleted chromosome 21 had no
yellow signal for the 21qter locus whereas has a signal for the 21q22 region. Here the
4qter were for they appeared on the same locus with 21q22 (Sans, Ezan, Moreau &
Montcouquiol, 2016).
(c) By use of the RUNX1/RUNX1TI dual color set of fusion translocation probe, the FISH
study demonstrated the RUNX1 signal locus.
(d) The array CGH detected the content rearrangement on the long arm of the chromosome
21 which had two copy number losses and three copy number gains.
(e) The other copy number variation indicated by the broken line in (d) composed of several
annotated genes of RUNX1.
The study was carried out to investigate the learning disability in a 17-year-old boy. The
array CGH is used to identify the interstitial deletion that involved a single clone 17q21.31
which will be confirmed by the Fish quantitative analysis (Hnoonual et al, 2017).
The figure 2; below shows the photos of the abnormalities that were analyzed in the above-
motioned study of a 17-year-old boy by the microarray at the tilling path resolution done to
investigate and refine the findings of the abnormalities (Pivalizza, & Seema, 2016).
(a) The fluorescence in situ hybridization (FISH) study involved and acro-p probe that
targeted short arms of the acrocentric chromosomes to investigate the additional satellite
on the long arm with 21qs. The (symbol *) showed that the G-banded 21qs chromosome
got an additional satellite on its long arm.
(b) The study showed that the 21q22 chromosome region composed of RUNXI gene and the
21qter locus aligned the deleted chromosomes. The deleted chromosome 21 had no
yellow signal for the 21qter locus whereas has a signal for the 21q22 region. Here the
4qter were for they appeared on the same locus with 21q22 (Sans, Ezan, Moreau &
Montcouquiol, 2016).
(c) By use of the RUNX1/RUNX1TI dual color set of fusion translocation probe, the FISH
study demonstrated the RUNX1 signal locus.
(d) The array CGH detected the content rearrangement on the long arm of the chromosome
21 which had two copy number losses and three copy number gains.
(e) The other copy number variation indicated by the broken line in (d) composed of several
annotated genes of RUNX1.
The study was carried out to investigate the learning disability in a 17-year-old boy. The
array CGH is used to identify the interstitial deletion that involved a single clone 17q21.31
which will be confirmed by the Fish quantitative analysis (Hnoonual et al, 2017).
The figure 2; below shows the photos of the abnormalities that were analyzed in the above-
motioned study of a 17-year-old boy by the microarray at the tilling path resolution done to
investigate and refine the findings of the abnormalities (Pivalizza, & Seema, 2016).
GENOMIC HEALTH 7
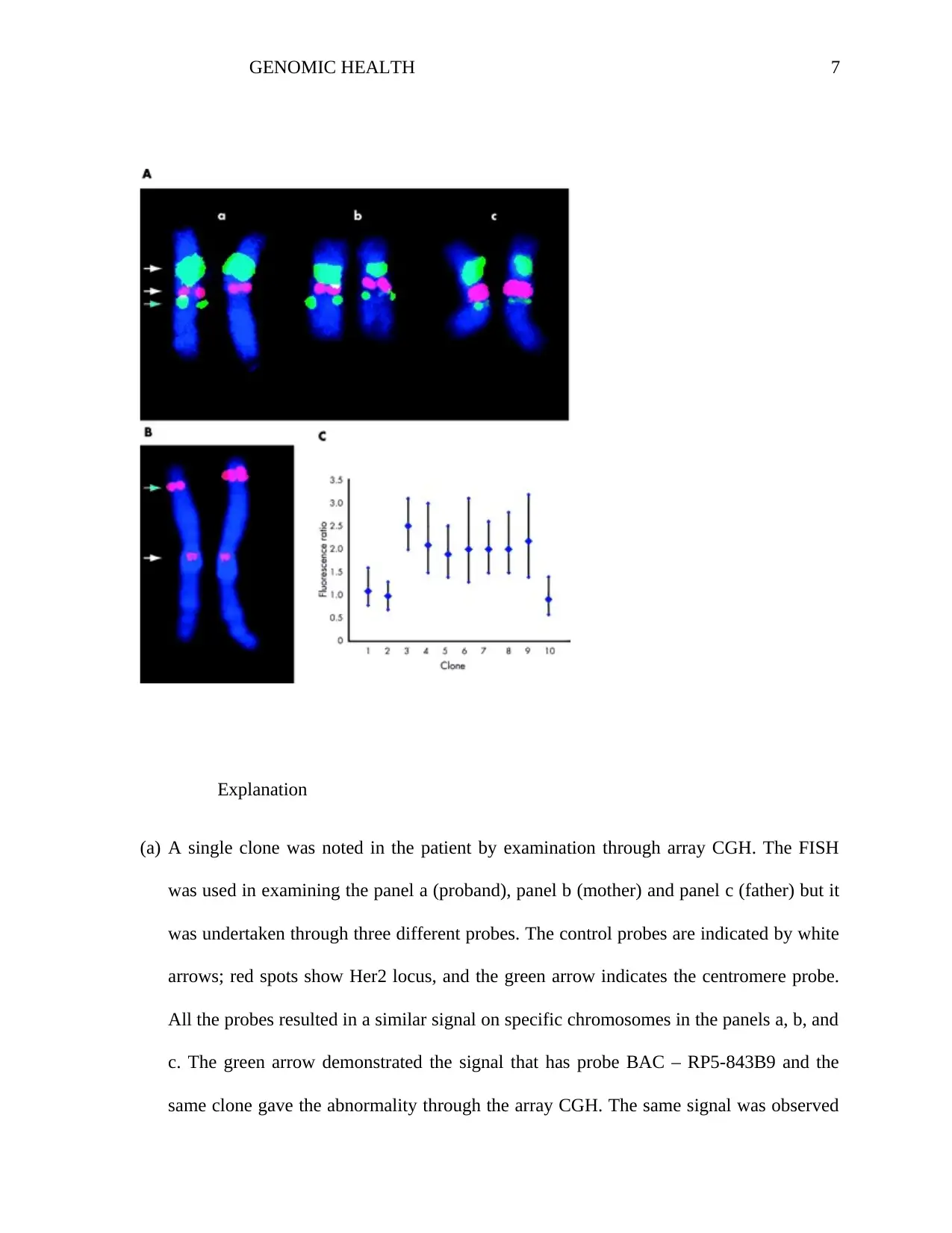
Explanation
(a) A single clone was noted in the patient by examination through array CGH. The FISH
was used in examining the panel a (proband), panel b (mother) and panel c (father) but it
was undertaken through three different probes. The control probes are indicated by white
arrows; red spots show Her2 locus, and the green arrow indicates the centromere probe.
All the probes resulted in a similar signal on specific chromosomes in the panels a, b, and
c. The green arrow demonstrated the signal that has probe BAC – RP5-843B9 and the
same clone gave the abnormality through the array CGH. The same signal was observed
Explanation
(a) A single clone was noted in the patient by examination through array CGH. The FISH
was used in examining the panel a (proband), panel b (mother) and panel c (father) but it
was undertaken through three different probes. The control probes are indicated by white
arrows; red spots show Her2 locus, and the green arrow indicates the centromere probe.
All the probes resulted in a similar signal on specific chromosomes in the panels a, b, and
c. The green arrow demonstrated the signal that has probe BAC – RP5-843B9 and the
same clone gave the abnormality through the array CGH. The same signal was observed
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

GENOMIC HEALTH 8
on the two chromosomes in panels c and b while on one chromosome was observed in
panel indicating the de novo deletion (Ocampos et al, 2017).
(b) The array CGH identified the duplications that involved five clones. The quantitative
FISH analysis was conducted on the proband by use of clones from the reputed
duplicated locus. It resulted in one chromosome observed. The noted differences in the
signal intensity and size between the normal chromosome 1 (left of the figure) and the
duplication chromosome 1 (right of the figure) which are observable through inspection.
The signal was quantified from both adjacent clones and within the duplicated putative
region.
(c) In the result of the FISH quantitative analysis, the fluorescence intensity of ratio2 is
consistency in putative duplication region while the ratio of 1 is consistency with no
duplication in the region outside which gave an intensity ratio of 1:1. The vertical bars
represent the 95% confidence limits. Through the analysis of the phenotype samples, it
was concluded that the duplication was due to the de novo event (Liehr, Glaser, &
Kosyakova, 2017).
Challenges in dealing with incidental findings
The new technology of array-CGH reports the findings that are highly likely to
explain the genomic symptoms and signs of the patient. However, some of the problems that
may arise are as follows;
The uncertainty significance of CNV
The main issue here is how to prepare both the patient and his family in advance about the
clinical significance uncertainty in the CNCs IT is not easy to explain to clients and his/her
on the two chromosomes in panels c and b while on one chromosome was observed in
panel indicating the de novo deletion (Ocampos et al, 2017).
(b) The array CGH identified the duplications that involved five clones. The quantitative
FISH analysis was conducted on the proband by use of clones from the reputed
duplicated locus. It resulted in one chromosome observed. The noted differences in the
signal intensity and size between the normal chromosome 1 (left of the figure) and the
duplication chromosome 1 (right of the figure) which are observable through inspection.
The signal was quantified from both adjacent clones and within the duplicated putative
region.
(c) In the result of the FISH quantitative analysis, the fluorescence intensity of ratio2 is
consistency in putative duplication region while the ratio of 1 is consistency with no
duplication in the region outside which gave an intensity ratio of 1:1. The vertical bars
represent the 95% confidence limits. Through the analysis of the phenotype samples, it
was concluded that the duplication was due to the de novo event (Liehr, Glaser, &
Kosyakova, 2017).
Challenges in dealing with incidental findings
The new technology of array-CGH reports the findings that are highly likely to
explain the genomic symptoms and signs of the patient. However, some of the problems that
may arise are as follows;
The uncertainty significance of CNV
The main issue here is how to prepare both the patient and his family in advance about the
clinical significance uncertainty in the CNCs IT is not easy to explain to clients and his/her

GENOMIC HEALTH 9
family about the genetic variation in humans. Due to the high risk of giving confronted findings
if the interpretation of the array- CGH was inaccurate, thus such findings uncertainty may cause
anxiety to the client and the family members (Williams III et al, 2015).
Variance in penetration of CNV
According to the array CGH, there are valuable numbers of neuro-susceptibility regions that
decrease the threshold for manifesting the autistic spectrum disorder, developmental delay, and
intellectual challenges. While investigating a child with the above symptoms, one of his/her
parents may be found to carry the same CNVS, but he/she has no history of such symptoms. This
is what is referred to as the reduced penetrance, and it is assumed that there are some additional
factors necessary to make the symptoms manifest.
Uncertain incidental findings
Sometimes the test may detect a clinical significance duplication or deletion which is not the
probable cause of the manifested symptoms for the patient; however, it may be having future
health implications in his/her generations or be within their relatives. An example of such an
incident is the deletion of cancer-predisposing gene. These are the feedback result that the patient
needs to be counseled about regarding the prophylactic surgery and such screening programmers
(Hanson et al, 2015).
Conclusion
In conclusion, the wide application of array-CCG in genomic examination has significantly
improved the identification of the rearrangement of the complex chromosomes and DNA copy
variance. Also, the technique has revolutionized the diagnosis result of patients with dysmorphic
family about the genetic variation in humans. Due to the high risk of giving confronted findings
if the interpretation of the array- CGH was inaccurate, thus such findings uncertainty may cause
anxiety to the client and the family members (Williams III et al, 2015).
Variance in penetration of CNV
According to the array CGH, there are valuable numbers of neuro-susceptibility regions that
decrease the threshold for manifesting the autistic spectrum disorder, developmental delay, and
intellectual challenges. While investigating a child with the above symptoms, one of his/her
parents may be found to carry the same CNVS, but he/she has no history of such symptoms. This
is what is referred to as the reduced penetrance, and it is assumed that there are some additional
factors necessary to make the symptoms manifest.
Uncertain incidental findings
Sometimes the test may detect a clinical significance duplication or deletion which is not the
probable cause of the manifested symptoms for the patient; however, it may be having future
health implications in his/her generations or be within their relatives. An example of such an
incident is the deletion of cancer-predisposing gene. These are the feedback result that the patient
needs to be counseled about regarding the prophylactic surgery and such screening programmers
(Hanson et al, 2015).
Conclusion
In conclusion, the wide application of array-CCG in genomic examination has significantly
improved the identification of the rearrangement of the complex chromosomes and DNA copy
variance. Also, the technique has revolutionized the diagnosis result of patients with dysmorphic

GENOMIC HEALTH 10
features, developmental delays, learning, and congenital anomalies. Most of the health
practitioners in the field of pediatrics and genetics use of array CGH screening tests enable them
to give clear information to the families with child’s difficulties such as learning disabilities,
developmental delay or any of the dysmorphic features. Although there may be no possible
curative treatment for such children’s difficulties, the genomic health specialist is able to give the
natural history of a particular family with the help of the microarray CGH analysis. Therefore the
findings can be of great assistance in devising therapeutic management and educational strategies
that may support children’s development and growth.
features, developmental delays, learning, and congenital anomalies. Most of the health
practitioners in the field of pediatrics and genetics use of array CGH screening tests enable them
to give clear information to the families with child’s difficulties such as learning disabilities,
developmental delay or any of the dysmorphic features. Although there may be no possible
curative treatment for such children’s difficulties, the genomic health specialist is able to give the
natural history of a particular family with the help of the microarray CGH analysis. Therefore the
findings can be of great assistance in devising therapeutic management and educational strategies
that may support children’s development and growth.
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

GENOMIC HEALTH 11
Reference
Durmaz, A. A., Karaca, E., Demkow, U., Toruner, G., Schoumans, J., & Cogulu, O. (2015).
Evolution of genetic techniques: past, present, and beyond. BioMed research
international, 2015.
Fragouli, E., Spath, K., Alfarawati, S., Kaper, F., Craig, A., Michel, C. E., ... & Wells, D. (2015).
Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and
provide an independent measure of embryonic implantation potential. PLoS genetics,
11(6), e1005241.
Giménez, C., Sarasa, J., Arjona, C., Vilamajó, E., Martínez-Pasarell, O., Wheeler, K., ... &
Wells, D. (2015). Karyomapping allows preimplantation genetic diagnosis of a de-novo
deletion undetectable using conventional PGD technology. Reproductive biomedicine
online, 31(6), 770-775.
Hanson, E., Bernier, R., Porche, K., Jackson, F. I., Goin-Kochel, R. P., Snyder, L. G., ... & Chen,
Q. (2015). The cognitive and behavioral phenotype of the 16p11. 2 deletion in a clinically
ascertained population. Biological psychiatry, 77(9), 785-793.
Hino-Fukuyo, N., Kikuchi, A., Arai-Ichinoi, N., Niihori, T., Sato, R., Suzuki, T., ... & Kubota, Y.
(2015). Genomic analysis identifies candidate pathogenic variants in 9 of 18 patients with
unexplained West syndrome. Human genetics, 134(6), 649-658.
Hnoonual, A., Thammachote, W., Tim-Aroon, T., Rojnueangnit, K., Hansakunachai, T.,
Sombuntham, T., ... & Wattanasirichaigoon, D. (2017). Chromosomal microarray
analysis in a cohort of underrepresented population identifies SERINC2 as a novel
candidate gene for autism spectrum disorder. Scientific reports, 7(1), 12096.
Reference
Durmaz, A. A., Karaca, E., Demkow, U., Toruner, G., Schoumans, J., & Cogulu, O. (2015).
Evolution of genetic techniques: past, present, and beyond. BioMed research
international, 2015.
Fragouli, E., Spath, K., Alfarawati, S., Kaper, F., Craig, A., Michel, C. E., ... & Wells, D. (2015).
Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and
provide an independent measure of embryonic implantation potential. PLoS genetics,
11(6), e1005241.
Giménez, C., Sarasa, J., Arjona, C., Vilamajó, E., Martínez-Pasarell, O., Wheeler, K., ... &
Wells, D. (2015). Karyomapping allows preimplantation genetic diagnosis of a de-novo
deletion undetectable using conventional PGD technology. Reproductive biomedicine
online, 31(6), 770-775.
Hanson, E., Bernier, R., Porche, K., Jackson, F. I., Goin-Kochel, R. P., Snyder, L. G., ... & Chen,
Q. (2015). The cognitive and behavioral phenotype of the 16p11. 2 deletion in a clinically
ascertained population. Biological psychiatry, 77(9), 785-793.
Hino-Fukuyo, N., Kikuchi, A., Arai-Ichinoi, N., Niihori, T., Sato, R., Suzuki, T., ... & Kubota, Y.
(2015). Genomic analysis identifies candidate pathogenic variants in 9 of 18 patients with
unexplained West syndrome. Human genetics, 134(6), 649-658.
Hnoonual, A., Thammachote, W., Tim-Aroon, T., Rojnueangnit, K., Hansakunachai, T.,
Sombuntham, T., ... & Wattanasirichaigoon, D. (2017). Chromosomal microarray
analysis in a cohort of underrepresented population identifies SERINC2 as a novel
candidate gene for autism spectrum disorder. Scientific reports, 7(1), 12096.

GENOMIC HEALTH 12
Kurtovic-Kozaric, A., Mehinovic, L., Stomornjak-Vukadin, M., Kurtovic-Basic, I., Catibusic, F.,
Kozaric, M., ... & Sumanovic-Glamuzina, D. (2016). Diagnostics of common
microdeletion syndromes using fluorescence in situ hybridization: single center
experience in a developing country. Bosnian journal of basic medical sciences, 16(2),
121.
Liehr, T., Glaser, A., & Kosyakova, N. (2017). Comparative Genomic Hybridization (CGH) and
Microdissection-Based CGH (Micro-CGH). In Fluorescence In Situ Hybridization
(FISH) (pp. 561-565). Springer, Berlin, Heidelberg.
Ocampos, M., Chaves, T. F., Barbato, I. T., de Luca, G. R., Barbato Filho, J. H., & Maris, A. F.
(2017). Partial Trisomy of 7q31. 32-q33, Revealed by Microarray CGH, in Three Patients
With Intellectual Disability, Hypotonia and Growth Delay. Cancer Genetics, 214, 48.
Pivalizza, P., & Seema, L. (2016). Intellectual disability in children: Evaluation for a cause.
UpToDate. Waltham, MA.
Sadek, A. A., & Mohamed, M. A. (2018). Yield of karyotyping in children with developmental
delay and/or dysmorphic features in Sohag University Hospital, Upper Egypt. Egyptian
Journal of Medical Human Genetics, 19(3), 253-259.
Sans, N., Ezan, J., Moreau, M. M., & Montcouquiol, M. (2016). Planar Cell Polarity Gene
Mutations in Autism Spectrum Disorder, Intellectual Disabilities, and Related
Deletion/Duplication Syndromes. In Neuronal and Synaptic Dysfunction in Autism
Spectrum Disorder and Intellectual Disability (pp. 189-219). Academic Press.
Rosenthal, E. T., Bernhisel, R., Brown, K., Kidd, J., & Manley, S. (2017). Clinical testing with a
panel of 25 genes associated with increased cancer risk results in a significant increase in
Kurtovic-Kozaric, A., Mehinovic, L., Stomornjak-Vukadin, M., Kurtovic-Basic, I., Catibusic, F.,
Kozaric, M., ... & Sumanovic-Glamuzina, D. (2016). Diagnostics of common
microdeletion syndromes using fluorescence in situ hybridization: single center
experience in a developing country. Bosnian journal of basic medical sciences, 16(2),
121.
Liehr, T., Glaser, A., & Kosyakova, N. (2017). Comparative Genomic Hybridization (CGH) and
Microdissection-Based CGH (Micro-CGH). In Fluorescence In Situ Hybridization
(FISH) (pp. 561-565). Springer, Berlin, Heidelberg.
Ocampos, M., Chaves, T. F., Barbato, I. T., de Luca, G. R., Barbato Filho, J. H., & Maris, A. F.
(2017). Partial Trisomy of 7q31. 32-q33, Revealed by Microarray CGH, in Three Patients
With Intellectual Disability, Hypotonia and Growth Delay. Cancer Genetics, 214, 48.
Pivalizza, P., & Seema, L. (2016). Intellectual disability in children: Evaluation for a cause.
UpToDate. Waltham, MA.
Sadek, A. A., & Mohamed, M. A. (2018). Yield of karyotyping in children with developmental
delay and/or dysmorphic features in Sohag University Hospital, Upper Egypt. Egyptian
Journal of Medical Human Genetics, 19(3), 253-259.
Sans, N., Ezan, J., Moreau, M. M., & Montcouquiol, M. (2016). Planar Cell Polarity Gene
Mutations in Autism Spectrum Disorder, Intellectual Disabilities, and Related
Deletion/Duplication Syndromes. In Neuronal and Synaptic Dysfunction in Autism
Spectrum Disorder and Intellectual Disability (pp. 189-219). Academic Press.
Rosenthal, E. T., Bernhisel, R., Brown, K., Kidd, J., & Manley, S. (2017). Clinical testing with a
panel of 25 genes associated with increased cancer risk results in a significant increase in

GENOMIC HEALTH 13
clinically significant findings across a broad range of cancer histories. Cancer genetics,
218, 58-68.
Williams III, J., Rad, S., Beauchamp, S., Ratousi, D., Subramaniam, V., Farivar, S., & Pisarska,
M. D. (2015). Utilization of noninvasive prenatal testing: impact on referrals for
diagnostic testing. American journal of obstetrics and gynecology, 213(1), 102-e1.
Xavier, J., Zhou, B., Bilan, F., Zhang, X., Gilbert-Dussardier, B., Viaux-Savelon, S., ... & Urban,
A. E. (2018). 1q21. 1 microduplication: large verbal–nonverbal performance discrepancy
and ddPCR assays of HYDIN/HYDIN2 copy number. NPJ genomic medicine, 3(1), 24.
clinically significant findings across a broad range of cancer histories. Cancer genetics,
218, 58-68.
Williams III, J., Rad, S., Beauchamp, S., Ratousi, D., Subramaniam, V., Farivar, S., & Pisarska,
M. D. (2015). Utilization of noninvasive prenatal testing: impact on referrals for
diagnostic testing. American journal of obstetrics and gynecology, 213(1), 102-e1.
Xavier, J., Zhou, B., Bilan, F., Zhang, X., Gilbert-Dussardier, B., Viaux-Savelon, S., ... & Urban,
A. E. (2018). 1q21. 1 microduplication: large verbal–nonverbal performance discrepancy
and ddPCR assays of HYDIN/HYDIN2 copy number. NPJ genomic medicine, 3(1), 24.
1 out of 13
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
© 2024 | Zucol Services PVT LTD | All rights reserved.