Reducing Drug Attrition: In Silico Modelling of Drug Toxicity
VerifiedAdded on 2022/08/22
|42
|7835
|13
Report
AI Summary
This report delves into the application of in silico modeling techniques to predict drug toxicity and mitigate drug attrition. It investigates the role of computational methods, such as DEREK Nexus software, in assessing the toxicity of drugs and the potential to reduce the failure rate of drugs in the market. The study focuses on methods like Quantitative Structure-Activity Relationship (QSAR) and Simplified Molecular Input Line Entry System (SMILES) notation to predict toxicities. The report also explores the use of Molecular Operating Environment (MOE) to generate 3D structures and SDF (Structure Data File) and examines how DEREK Nexus software can read these structures to predict toxicities. The results of the toxicity predictions are compared with historical data of withdrawn drugs to determine the effectiveness of the software. The report further discusses the role of in silico techniques in reducing drug attrition by identifying gaps in current safety screening cascades and minimizing animal testing. The report explores the significance of ADME (Absorption, Distribution, Metabolism, and Excretion) properties and the use of regression analysis in determining drug toxicity levels. The findings highlight the advantages of in silico modeling in predicting drug toxicity and reducing drug attrition during drug development.

In Silico Modelling of Drug Toxicity: Reducing Drug
Attrition
Attrition
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Content
s
Abbreviations:...................................................................................5
Abstract:............................................................................................6
Introduction:......................................................................................8
Role of In Silico Technique in Reducing Drug
Attrition……………………………….12
Figure 1: In Silico Technique Helps in Predicting Toxicites............................13
Figure 2: Steps Invoved in the Generation of Prediction
Models……………....16
Table 1:Example of Descriptors used In Silico Methods................................21
Methods:..........................................................................................23
Results:.............................................................................................25
Figure 3: Graph Shows Percetange of Agreement using DEREK Software....25
Table 2: Percentage of Predicted Drugs Adverse Reaction...........................25
Discussion: ......................................................................................27
Figure 4: Metabolic Oxidation of Nefazodone
Drug...................................27
Conculsion:......................................................................................32
References:……………………………………………………………………………….…
33
Appendices: ....................................................................................37
s
Abbreviations:...................................................................................5
Abstract:............................................................................................6
Introduction:......................................................................................8
Role of In Silico Technique in Reducing Drug
Attrition……………………………….12
Figure 1: In Silico Technique Helps in Predicting Toxicites............................13
Figure 2: Steps Invoved in the Generation of Prediction
Models……………....16
Table 1:Example of Descriptors used In Silico Methods................................21
Methods:..........................................................................................23
Results:.............................................................................................25
Figure 3: Graph Shows Percetange of Agreement using DEREK Software....25
Table 2: Percentage of Predicted Drugs Adverse Reaction...........................25
Discussion: ......................................................................................27
Figure 4: Metabolic Oxidation of Nefazodone
Drug...................................27
Conculsion:......................................................................................32
References:……………………………………………………………………………….…
33
Appendices: ....................................................................................37

Abbreviations:
ADME……Absorption, Distribution, Metabolism, and Excretion
MOE…..……………………Molecular Operating Environment
PK............................... Pharmacokinetics
QSAR………………Quantitative Structure-Activity Relationship
QSPR…………Quantity Structure Toxicity/Property Relationship
SMILES……….. Simplified Molecular Input Line Entry System
ADME……Absorption, Distribution, Metabolism, and Excretion
MOE…..……………………Molecular Operating Environment
PK............................... Pharmacokinetics
QSAR………………Quantitative Structure-Activity Relationship
QSPR…………Quantity Structure Toxicity/Property Relationship
SMILES……….. Simplified Molecular Input Line Entry System
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Abstract:
There is a greater need for most pharmaceutical industries to apply different
mechanism to address the issues of drug failure. Both pharmacokinetic and
toxicokinetic profiles play imperative roles in the drug discovery and
development. In silico methods is one of the alternatives to animal testing
that complement in vitro and in vivo toxicity assessments to potentially
minimise animal testing, reduce the cost and time of drug discovery, and
improve toxicity prediction and safety assessment.
This project is looking to see if modern software such as DEREK Nexus is
able to predict toxicities of drugs which have historically been withdrawn
from the market. The project also determines if the ability to use DEREK
software for toxicity prediction would have made a difference if they could
be available at the drugs discovery stage. SMILES notation for example has
been considered by most researchers as the most favourable method of
coding for chemical structures. The study employed the use of Molecular
Operating Environment (MOE) to generate 3D structures and the SDF
(Structure Data File). DEREK nexus software was then used to read the 3D
There is a greater need for most pharmaceutical industries to apply different
mechanism to address the issues of drug failure. Both pharmacokinetic and
toxicokinetic profiles play imperative roles in the drug discovery and
development. In silico methods is one of the alternatives to animal testing
that complement in vitro and in vivo toxicity assessments to potentially
minimise animal testing, reduce the cost and time of drug discovery, and
improve toxicity prediction and safety assessment.
This project is looking to see if modern software such as DEREK Nexus is
able to predict toxicities of drugs which have historically been withdrawn
from the market. The project also determines if the ability to use DEREK
software for toxicity prediction would have made a difference if they could
be available at the drugs discovery stage. SMILES notation for example has
been considered by most researchers as the most favourable method of
coding for chemical structures. The study employed the use of Molecular
Operating Environment (MOE) to generate 3D structures and the SDF
(Structure Data File). DEREK nexus software was then used to read the 3D
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

structures and the SDF generated by theme to predict toxicities. Based on
the data obtained from toxicity predictions by the DEREK Nexus software,
most of its toxicity predictions agree with the previously predicted toxicity
that leads to their withdrawal from the market. Few predictions were in
disagreement with the DEREK Nexus predictions. The use DEREK software
is effective for toxicity prediction. The use of structural alerts in Derek
nexus software in the identification of the reasons for drug withdrawal show
how effective the use expert systems are.
Introduction:
The discovery of various drugs requires a significant investment in
terms of time and resources. It has been considered very significant to
address the high attrition rates in drug development (Agamah et al, 2019).
Drug attrition is the failure of drugs to meet the market entry requirements
due to its adverse effects to the target. This is caused by high level of
toxicity during drug development stages. Toxicity can be defined as the
processing of measuring the side effects of drugs before released into the
clinics and further into the market (Berg 2019). The pharmaceutical
industries, therefore, remain under huge pressure to provide solutions to the
high rates of attrition in drug development.
the data obtained from toxicity predictions by the DEREK Nexus software,
most of its toxicity predictions agree with the previously predicted toxicity
that leads to their withdrawal from the market. Few predictions were in
disagreement with the DEREK Nexus predictions. The use DEREK software
is effective for toxicity prediction. The use of structural alerts in Derek
nexus software in the identification of the reasons for drug withdrawal show
how effective the use expert systems are.
Introduction:
The discovery of various drugs requires a significant investment in
terms of time and resources. It has been considered very significant to
address the high attrition rates in drug development (Agamah et al, 2019).
Drug attrition is the failure of drugs to meet the market entry requirements
due to its adverse effects to the target. This is caused by high level of
toxicity during drug development stages. Toxicity can be defined as the
processing of measuring the side effects of drugs before released into the
clinics and further into the market (Berg 2019). The pharmaceutical
industries, therefore, remain under huge pressure to provide solutions to the
high rates of attrition in drug development.

It is imperative to note that failure of drugs during early stages of
development attrition drug candidates remain a problem. This is despite the
continued application of various software such as TOPKAT, CASE,
ToxTree, and DEREK to predict toxicity from the structure of chemical/drug
compounds. Moreover, for drug development to meet both financial and
economic sustainability there is a great need to reduce animal testing for
toxicological risk assessment. The use of animal in testing for toxicity is
uneconomical in terms finance and time due to high chances of drug failing
to meet the requirements (Sakti, Nishimura, and Nakai, 2018). In silico can
be defined as the process of integration of modern computing and
information technology to improve the process of assessing drugs (Baur et
al., 2020). In silico methods, therefore, offer an alternative that can be used
to address the challenges associated with the high attrition rates in drug
development.
Poor pharmacokinetic and toxicokinetic profiles in the field of medicine
have significantly increased the number of drug failing at the late stage of
development. Toxicokinetic or biokinetics in toxicology refers to processes
such as absorption, distribution, metabolism, and excretion. The
pharmacokinetic profile of drugs that interact with living organisms
significantly relates to the above processes (Clippinger et al, 2018).The fate
development attrition drug candidates remain a problem. This is despite the
continued application of various software such as TOPKAT, CASE,
ToxTree, and DEREK to predict toxicity from the structure of chemical/drug
compounds. Moreover, for drug development to meet both financial and
economic sustainability there is a great need to reduce animal testing for
toxicological risk assessment. The use of animal in testing for toxicity is
uneconomical in terms finance and time due to high chances of drug failing
to meet the requirements (Sakti, Nishimura, and Nakai, 2018). In silico can
be defined as the process of integration of modern computing and
information technology to improve the process of assessing drugs (Baur et
al., 2020). In silico methods, therefore, offer an alternative that can be used
to address the challenges associated with the high attrition rates in drug
development.
Poor pharmacokinetic and toxicokinetic profiles in the field of medicine
have significantly increased the number of drug failing at the late stage of
development. Toxicokinetic or biokinetics in toxicology refers to processes
such as absorption, distribution, metabolism, and excretion. The
pharmacokinetic profile of drugs that interact with living organisms
significantly relates to the above processes (Clippinger et al, 2018).The fate
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

of any drug inside the body of an organism is determined by the four
processes.
The overall profiling properties of the four processes and toxic effects
of substances is expressed by the term ADME (T). In silico methods
significantly rely on the ADME properties of compounds to test and assess
the side effects of any drug before they are released for use. The ADME (T)
properties of compounds include absorption, distribution, metabolism and
excretion properties used especially by the in silico methods to test for drug
toxicity level. In silico methods give a better interpretation of in silico
toxicity data for their relevance in terms of a toxic dose by incorporating the
use of physiologically based biokinetics modelling. The concentration of a
given dose in a tissue is usually determined by this model (Naven, & Louise-
May, 2015). It also estimates the effective concentration of an external dose
in the target tissue. In integrated testing work in unison within in silico
methods to make predictions on chemical compounds.
The modelling of various physiological properties of drugs such as
toxicity, metabolism, drug-drug interactions, and carcinogenesis have been
facilitated by the use of Quantitative Structure-Activity Relationship
(QSAR) techniques. The application and use of QSAR in modelling various
physiological properties of drugs require the application of SMILES
processes.
The overall profiling properties of the four processes and toxic effects
of substances is expressed by the term ADME (T). In silico methods
significantly rely on the ADME properties of compounds to test and assess
the side effects of any drug before they are released for use. The ADME (T)
properties of compounds include absorption, distribution, metabolism and
excretion properties used especially by the in silico methods to test for drug
toxicity level. In silico methods give a better interpretation of in silico
toxicity data for their relevance in terms of a toxic dose by incorporating the
use of physiologically based biokinetics modelling. The concentration of a
given dose in a tissue is usually determined by this model (Naven, & Louise-
May, 2015). It also estimates the effective concentration of an external dose
in the target tissue. In integrated testing work in unison within in silico
methods to make predictions on chemical compounds.
The modelling of various physiological properties of drugs such as
toxicity, metabolism, drug-drug interactions, and carcinogenesis have been
facilitated by the use of Quantitative Structure-Activity Relationship
(QSAR) techniques. The application and use of QSAR in modelling various
physiological properties of drugs require the application of SMILES
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

(Simplified Molecular Input Line Entry System). SMILES is a scientific
language usually developed to process information on modern chemistry
(Rognan, 2017). Moreover, this technique applies the principles of
molecular graph theory and very small and natural grammar to allow
rigorous structure specification. Molecular graph theory is the use of graph
to represent the structural formula of a chemical compound.
Most of the highly efficient chemical computer applications are
designed to generate unique chemical notation to allow constant-speed
database retrieval, flexible substructure searching, and property prediction
models through their ease of usage by the chemist and machine
compatibility. SMILES is a language used in software to code for structure.
The use and application of SMILES in predicting toxicities are due to
several advantages. Due to its ease of use and accessibility by chemists,
interpretation and generation of chemical notation that are independent of
the specific computer system in use is significantly simplified (Przekwas and
Somayaji, 2018). The ability of SMILES to represent molecular structure
using a linear string of symbols similar to natural language makes is use
very popular by most computational chemist in predicting drug toxicity.
SMILES notation simplifies the chemical structure of chemical compounds
through a two-dimensional valence-oriented picture graph. Hydrogen can
language usually developed to process information on modern chemistry
(Rognan, 2017). Moreover, this technique applies the principles of
molecular graph theory and very small and natural grammar to allow
rigorous structure specification. Molecular graph theory is the use of graph
to represent the structural formula of a chemical compound.
Most of the highly efficient chemical computer applications are
designed to generate unique chemical notation to allow constant-speed
database retrieval, flexible substructure searching, and property prediction
models through their ease of usage by the chemist and machine
compatibility. SMILES is a language used in software to code for structure.
The use and application of SMILES in predicting toxicities are due to
several advantages. Due to its ease of use and accessibility by chemists,
interpretation and generation of chemical notation that are independent of
the specific computer system in use is significantly simplified (Przekwas and
Somayaji, 2018). The ability of SMILES to represent molecular structure
using a linear string of symbols similar to natural language makes is use
very popular by most computational chemist in predicting drug toxicity.
SMILES notation simplifies the chemical structure of chemical compounds
through a two-dimensional valence-oriented picture graph. Hydrogen can

either be included or excluded in the representation of molecular structure
using this language (Worth, 2018). The use of SMILES follows certain rules
and specifications. Letters are always used to represent the atoms within the
structure as shown below.
CC Ethane (C2H6)
N ammine (NH2)
For toxicity of various drugs to be predictd by Derek software, structure
must first be encoded in the SMILES language.
is for example encoded in the SMILE language as
O=C-O-C-C-C
SMILES notation has been considered by most researchers as the most
favourable method for developing a large database for molecular structures
(Effinger et al, 2018. QSAR is a technique that analyses the equation
connecting the structures of molecules to their respective measured activity
using this language (Worth, 2018). The use of SMILES follows certain rules
and specifications. Letters are always used to represent the atoms within the
structure as shown below.
CC Ethane (C2H6)
N ammine (NH2)
For toxicity of various drugs to be predictd by Derek software, structure
must first be encoded in the SMILES language.
is for example encoded in the SMILE language as
O=C-O-C-C-C
SMILES notation has been considered by most researchers as the most
favourable method for developing a large database for molecular structures
(Effinger et al, 2018. QSAR is a technique that analyses the equation
connecting the structures of molecules to their respective measured activity
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

and property to predict the activity, reactivity, and properties of an unknown
set of molecules. As earlier mentioned, there is a great need to provide
solutions to the problem drug attrition in both early and late stages of drug
development as well as the need to reduce animal testing for toxicological
risk assessment. This project, therefore, investigates the capabilities of the
modern software in predicting toxicities of drugs which have historically
been withdrawn from the market.
Role of in silico technique in reducing drug attrition:
In silico technique has been extensively applied to help in reducing
the rate at which new drugs are failing in pharmaceutical research and
development. This is due to its ability to identify the gaps that exist in the
current mechanistic and chemical toxicology used in the testing of drug
toxicity levels (Atienzar et al 2016). Computational toxicology has,
therefore, engaged in understanding these gaps to provide the solution to
factors that lead to high drug attrition rate. This is through the application of
modern software such DEREK nexus in enhancing toxicity prediction. In
silico models tend to provide solutions to drug failure by identifying the
gaps that exist in the current safety screening cascades through
set of molecules. As earlier mentioned, there is a great need to provide
solutions to the problem drug attrition in both early and late stages of drug
development as well as the need to reduce animal testing for toxicological
risk assessment. This project, therefore, investigates the capabilities of the
modern software in predicting toxicities of drugs which have historically
been withdrawn from the market.
Role of in silico technique in reducing drug attrition:
In silico technique has been extensively applied to help in reducing
the rate at which new drugs are failing in pharmaceutical research and
development. This is due to its ability to identify the gaps that exist in the
current mechanistic and chemical toxicology used in the testing of drug
toxicity levels (Atienzar et al 2016). Computational toxicology has,
therefore, engaged in understanding these gaps to provide the solution to
factors that lead to high drug attrition rate. This is through the application of
modern software such DEREK nexus in enhancing toxicity prediction. In
silico models tend to provide solutions to drug failure by identifying the
gaps that exist in the current safety screening cascades through
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

understanding the structural, mechanistic, chemical, and the
pharmacological space of the drug molecules (Ekins et al, 2019).
The model contributes significantly in minimizing the rate of drug
failure by identifying the toxicological knowledge gaps that exist in the
already exposed miss-predicted compounds through validated analytical
processes that provide the means for understanding the applicability domain
of each assay or model. The applicability domain (AD) of a QSAR model is
the physico-chemical, structural or biological space, knowledge or
information on which the training set of the model has been developed, and
for which it is applicable to make predictions for new compounds. Naven &
Louise-May conducted a study using regression analysis to analyse
preclinical drugs that were annotated concerning their level of toxicity. Their
findings indicate that in vivo are not very effective in predicting toxicities of
compounds (Worth, 2018). The use of regression analysis yielded an
increased number of drug candidates in the preclinical screening that could
be rendered toxic if the in vivo technique could be used. Therefore, in vivo
technique is ineffective predicting drug toxicity. This was due to the ability
of this method to predict its toxicity level based on the drug physicochemical
properties, conclusively, in silico modeling method is essential in reducing
drug attrition during drug development.
pharmacological space of the drug molecules (Ekins et al, 2019).
The model contributes significantly in minimizing the rate of drug
failure by identifying the toxicological knowledge gaps that exist in the
already exposed miss-predicted compounds through validated analytical
processes that provide the means for understanding the applicability domain
of each assay or model. The applicability domain (AD) of a QSAR model is
the physico-chemical, structural or biological space, knowledge or
information on which the training set of the model has been developed, and
for which it is applicable to make predictions for new compounds. Naven &
Louise-May conducted a study using regression analysis to analyse
preclinical drugs that were annotated concerning their level of toxicity. Their
findings indicate that in vivo are not very effective in predicting toxicities of
compounds (Worth, 2018). The use of regression analysis yielded an
increased number of drug candidates in the preclinical screening that could
be rendered toxic if the in vivo technique could be used. Therefore, in vivo
technique is ineffective predicting drug toxicity. This was due to the ability
of this method to predict its toxicity level based on the drug physicochemical
properties, conclusively, in silico modeling method is essential in reducing
drug attrition during drug development.
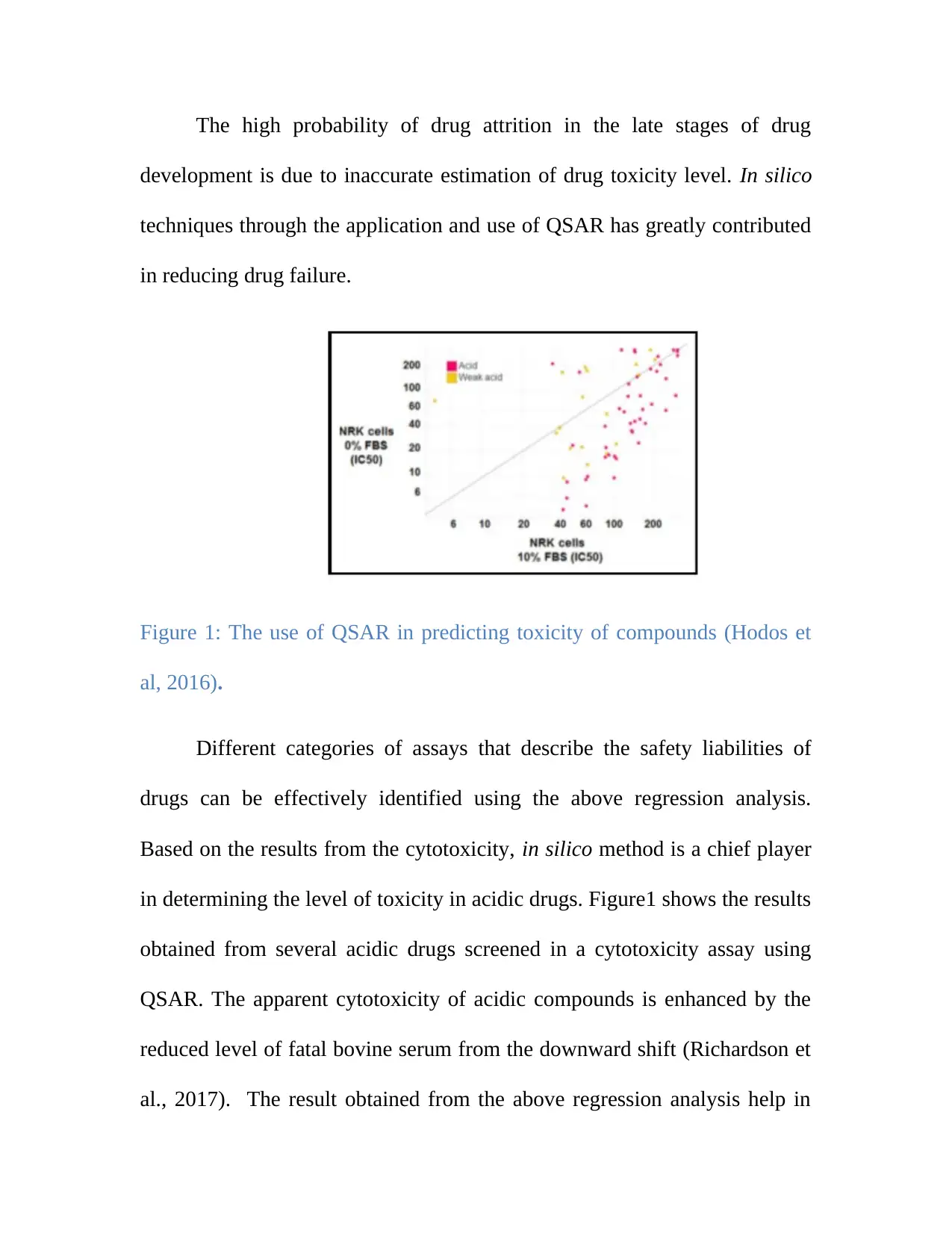
The high probability of drug attrition in the late stages of drug
development is due to inaccurate estimation of drug toxicity level. In silico
techniques through the application and use of QSAR has greatly contributed
in reducing drug failure.
Figure 1: The use of QSAR in predicting toxicity of compounds (Hodos et
al, 2016).
Different categories of assays that describe the safety liabilities of
drugs can be effectively identified using the above regression analysis.
Based on the results from the cytotoxicity, in silico method is a chief player
in determining the level of toxicity in acidic drugs. Figure1 shows the results
obtained from several acidic drugs screened in a cytotoxicity assay using
QSAR. The apparent cytotoxicity of acidic compounds is enhanced by the
reduced level of fatal bovine serum from the downward shift (Richardson et
al., 2017). The result obtained from the above regression analysis help in
development is due to inaccurate estimation of drug toxicity level. In silico
techniques through the application and use of QSAR has greatly contributed
in reducing drug failure.
Figure 1: The use of QSAR in predicting toxicity of compounds (Hodos et
al, 2016).
Different categories of assays that describe the safety liabilities of
drugs can be effectively identified using the above regression analysis.
Based on the results from the cytotoxicity, in silico method is a chief player
in determining the level of toxicity in acidic drugs. Figure1 shows the results
obtained from several acidic drugs screened in a cytotoxicity assay using
QSAR. The apparent cytotoxicity of acidic compounds is enhanced by the
reduced level of fatal bovine serum from the downward shift (Richardson et
al., 2017). The result obtained from the above regression analysis help in
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 42
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2025 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.
