Morphological & Biochemical Characterization of Unknown Bacteria B
VerifiedAdded on 2023/06/04
|6
|2023
|228
Practical Assignment
AI Summary
This assignment focuses on the isolation and characterization of unknown bacteria B through a series of morphological and biochemical tests. The experiment begins with a physical examination of colony characteristics on nutrient agar, followed by Gram staining to determine cell morphology. Motility tests are conducted to assess bacterial movement, and multiple biochemical tests, including Indole, Catalase, Starch hydrolysis, and Gelatin hydrolysis, are performed to identify specific enzymatic capabilities. Oxygen utilization and sugar fermentation tests further classify the bacteria's metabolic properties. The results of these tests are compiled to identify the unknown bacteria, ultimately concluding that bacteria B is Bacillus subtilis, based on its Gram-positive, rod-shaped morphology, motility, aerobic nature, and specific biochemical reactions. The conclusion highlights the medical importance of Bacillus subtilis and its probiotic characteristics.

Introduction
In microbiology isolation and characterization of microorganism is important. This is critical in
understanding particular disease etiology and the best antibiotic to be used for the diseases
treatment and the abilities of the bacteria to produce substances that might help it evade
treatment such as having antibiotic resistance capabilities.
Aim of the laboratory practical
The aim of this laboratory experiment was to isolate and characterize the unknown bacteria B
using morphological and biochemical tests. Characterization tests included gram staining,
microscopy to check for cellular morphology, spore formation, motility, Indole test, fermentation
ability, amylase, catalase, and gelatinase production.
Materials and Methods
Physical examination of the colony characteristics on nutrient agar was the first step to be
performed before gram staining. A single colony of unknown bacteria B was picked using sterile
wire loop and spread on glass slide. The crystal violet was used as the primary stain, 95% ethyl
alcohol was used to decolorize the primary stain, while Gram’s iodine was used as a mordant,
and safranin used as counter stain. The Gram positive bacteria retain the primary color which is
dark purple (Pogmore, Seistrup, & Strahl, 2018). The unknown bacteria B stained deed purple
when observed under microscope with rod shape.
Motility test was carried out to determine if the unknown bacteria (B) was motile or non-motile.
The motility of the bacteria B was established by stab inoculation of the single colony of the
bacteria B into the motility media tubes provided. The presence of diffuse growth away from the
stab line of inoculation, demonstrated by turbidity extending sideways across the motility test
medium was an indicator for positive test (Luna et al., 2005). Presence of growth confined to the
stab line was indicated a negative motility test.
Unknown bacterial B was further identified using multiple biochemical tests. Bacteria B was
tested further to identify its ability to form spores and the type of spore produced if any. Based
on the result interpretation from the manual there was no spore formation. Indole test was
conducted to establish the ability of the unknown bacteria B to produce enzyme tryptophanase
that breaks down tryptophan and reacts with Kovac’s reagent. According to Abbott (2011),
1
In microbiology isolation and characterization of microorganism is important. This is critical in
understanding particular disease etiology and the best antibiotic to be used for the diseases
treatment and the abilities of the bacteria to produce substances that might help it evade
treatment such as having antibiotic resistance capabilities.
Aim of the laboratory practical
The aim of this laboratory experiment was to isolate and characterize the unknown bacteria B
using morphological and biochemical tests. Characterization tests included gram staining,
microscopy to check for cellular morphology, spore formation, motility, Indole test, fermentation
ability, amylase, catalase, and gelatinase production.
Materials and Methods
Physical examination of the colony characteristics on nutrient agar was the first step to be
performed before gram staining. A single colony of unknown bacteria B was picked using sterile
wire loop and spread on glass slide. The crystal violet was used as the primary stain, 95% ethyl
alcohol was used to decolorize the primary stain, while Gram’s iodine was used as a mordant,
and safranin used as counter stain. The Gram positive bacteria retain the primary color which is
dark purple (Pogmore, Seistrup, & Strahl, 2018). The unknown bacteria B stained deed purple
when observed under microscope with rod shape.
Motility test was carried out to determine if the unknown bacteria (B) was motile or non-motile.
The motility of the bacteria B was established by stab inoculation of the single colony of the
bacteria B into the motility media tubes provided. The presence of diffuse growth away from the
stab line of inoculation, demonstrated by turbidity extending sideways across the motility test
medium was an indicator for positive test (Luna et al., 2005). Presence of growth confined to the
stab line was indicated a negative motility test.
Unknown bacterial B was further identified using multiple biochemical tests. Bacteria B was
tested further to identify its ability to form spores and the type of spore produced if any. Based
on the result interpretation from the manual there was no spore formation. Indole test was
conducted to establish the ability of the unknown bacteria B to produce enzyme tryptophanase
that breaks down tryptophan and reacts with Kovac’s reagent. According to Abbott (2011),
1
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Indole positive result is indicated by red surface layer while negative result doesn’t have the red
surface layer.
Catalase test was also performed. Catalase test is important in differentiating catalase producing
bacteria from non-catalase producing bacteria. In this test catalase acts by catalyzing the
breakdown of hydrogen peroxide to oxygen and water. The bubbles of oxygen are produced in
the presence of catalase producing bacteria while catalase negative bacteria don’t produce
bubbles in presence of hydrogen peroxide (Cheesbrough, 2006).
Starch hydrolysis test was carried out on unknown bacteria B to check its ability to produce
amylase that can hydrolyze starch. This test involved the use of a differential media which was
starch agar plate. The media was inoculated by Unknown bacteria B. The starch agar plate with
colonies of bacteria B was then flooded with Gram’s iodine. Clearing of light yellow/gold zone
around bacterial colonies indicated presence of amylase while no clearing around the colonies
indicated a negative amylase results (Xia et al., 2015). From the result there was clearing of light
yellow zone indicating presence of amylase.
The ability of unknown bacteria B to produce gelatinase which liquefy gelatin was tested by
performing gelatin hydrolysis test. The hydrolysis process by the gelatinase enzyme takes place
in two sequential reaction. In the first reaction, the enzyme gelatinase breakdown the gelatin to
polypeptide. Polypeptide is then further degraded to amino acid (Cheesbrough, 2006). A positive
result was indicated by a partial liquefaction of the inoculated tube even after being subjected ice
water.
Oxygen utilization test was performed to determine whether the unknown bacteria B was aerobic
or anaerobic bacteria. Thioglycollate broth which is an enrichment media was used, the media
contain numerous nutrient factors such as casein, yeast, beef extract, and oxidation-reduction
indictor (resazurin). It supports growth of anaerobes, aerobes, microaerophilic, and fastidious
microorganisms. The growth in the upper part of the media where the oxygen concentration was
high is indicative of aerobic bacteria while anaerobic bacteria grows to the bottom of the media
(Cowan, 2005)
The ability of the bacteria B to ferment lactose and glucose was tested. Phenol Red Lactose
Broth, Phenol Red Glucose Broth were used. Sugar fermenting bacteria produces acid during
fermentation that lowers the PH of the broth, hence, the phenol red indicator which is red in
colour will change its colour to yellow due to acid production. At the same time, the gas
produced is collected inside Durham’s tube. Therefore, a positive sugar fermentation is indicated
by change in the colour of the broth from red to yellow (Cowan, 2005).
2
surface layer.
Catalase test was also performed. Catalase test is important in differentiating catalase producing
bacteria from non-catalase producing bacteria. In this test catalase acts by catalyzing the
breakdown of hydrogen peroxide to oxygen and water. The bubbles of oxygen are produced in
the presence of catalase producing bacteria while catalase negative bacteria don’t produce
bubbles in presence of hydrogen peroxide (Cheesbrough, 2006).
Starch hydrolysis test was carried out on unknown bacteria B to check its ability to produce
amylase that can hydrolyze starch. This test involved the use of a differential media which was
starch agar plate. The media was inoculated by Unknown bacteria B. The starch agar plate with
colonies of bacteria B was then flooded with Gram’s iodine. Clearing of light yellow/gold zone
around bacterial colonies indicated presence of amylase while no clearing around the colonies
indicated a negative amylase results (Xia et al., 2015). From the result there was clearing of light
yellow zone indicating presence of amylase.
The ability of unknown bacteria B to produce gelatinase which liquefy gelatin was tested by
performing gelatin hydrolysis test. The hydrolysis process by the gelatinase enzyme takes place
in two sequential reaction. In the first reaction, the enzyme gelatinase breakdown the gelatin to
polypeptide. Polypeptide is then further degraded to amino acid (Cheesbrough, 2006). A positive
result was indicated by a partial liquefaction of the inoculated tube even after being subjected ice
water.
Oxygen utilization test was performed to determine whether the unknown bacteria B was aerobic
or anaerobic bacteria. Thioglycollate broth which is an enrichment media was used, the media
contain numerous nutrient factors such as casein, yeast, beef extract, and oxidation-reduction
indictor (resazurin). It supports growth of anaerobes, aerobes, microaerophilic, and fastidious
microorganisms. The growth in the upper part of the media where the oxygen concentration was
high is indicative of aerobic bacteria while anaerobic bacteria grows to the bottom of the media
(Cowan, 2005)
The ability of the bacteria B to ferment lactose and glucose was tested. Phenol Red Lactose
Broth, Phenol Red Glucose Broth were used. Sugar fermenting bacteria produces acid during
fermentation that lowers the PH of the broth, hence, the phenol red indicator which is red in
colour will change its colour to yellow due to acid production. At the same time, the gas
produced is collected inside Durham’s tube. Therefore, a positive sugar fermentation is indicated
by change in the colour of the broth from red to yellow (Cowan, 2005).
2
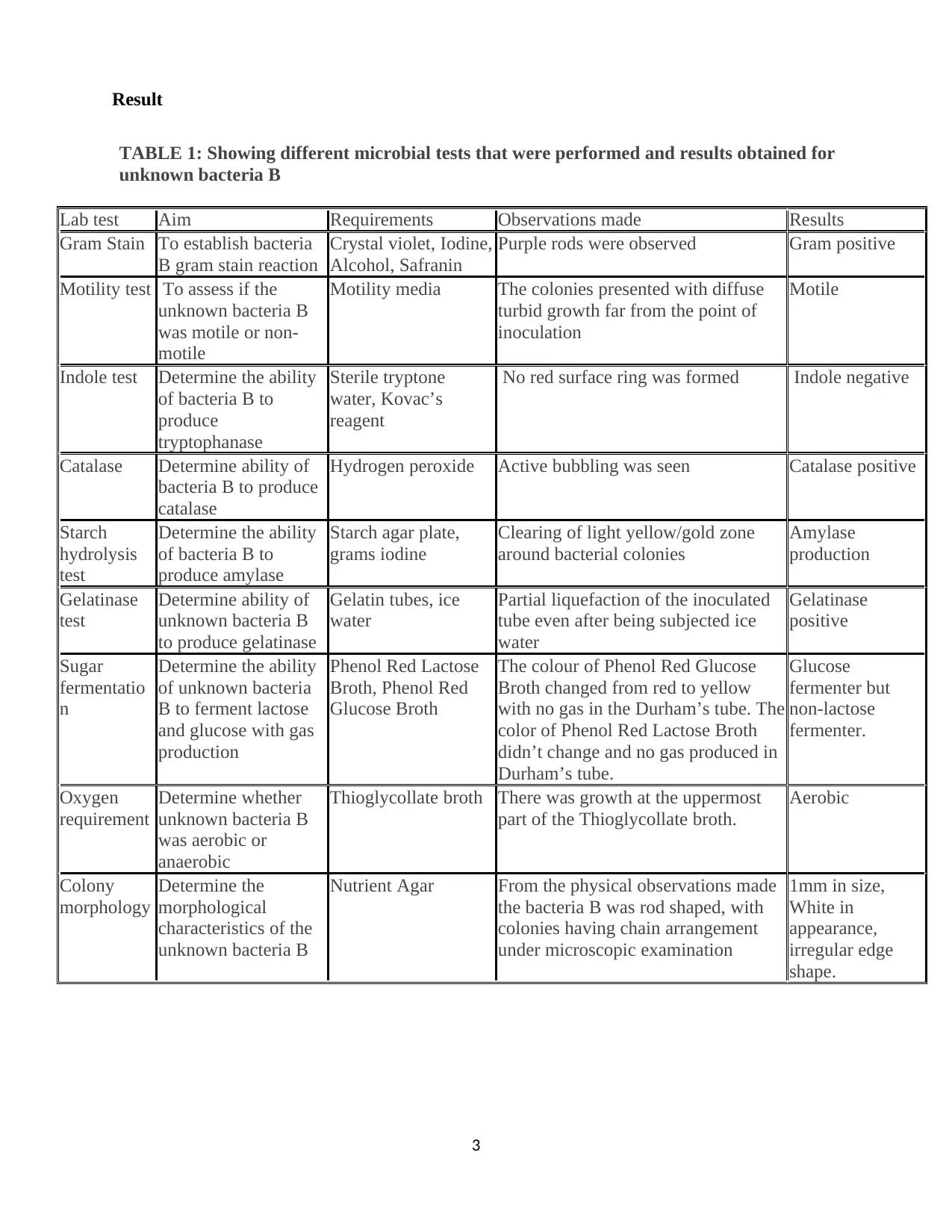
Result
TABLE 1: Showing different microbial tests that were performed and results obtained for
unknown bacteria B
Lab test Aim Requirements Observations made Results
Gram Stain To establish bacteria
B gram stain reaction
Crystal violet, Iodine,
Alcohol, Safranin
Purple rods were observed Gram positive
Motility test To assess if the
unknown bacteria B
was motile or non-
motile
Motility media The colonies presented with diffuse
turbid growth far from the point of
inoculation
Motile
Indole test Determine the ability
of bacteria B to
produce
tryptophanase
Sterile tryptone
water, Kovac’s
reagent
No red surface ring was formed Indole negative
Catalase Determine ability of
bacteria B to produce
catalase
Hydrogen peroxide Active bubbling was seen Catalase positive
Starch
hydrolysis
test
Determine the ability
of bacteria B to
produce amylase
Starch agar plate,
grams iodine
Clearing of light yellow/gold zone
around bacterial colonies
Amylase
production
Gelatinase
test
Determine ability of
unknown bacteria B
to produce gelatinase
Gelatin tubes, ice
water
Partial liquefaction of the inoculated
tube even after being subjected ice
water
Gelatinase
positive
Sugar
fermentatio
n
Determine the ability
of unknown bacteria
B to ferment lactose
and glucose with gas
production
Phenol Red Lactose
Broth, Phenol Red
Glucose Broth
The colour of Phenol Red Glucose
Broth changed from red to yellow
with no gas in the Durham’s tube. The
color of Phenol Red Lactose Broth
didn’t change and no gas produced in
Durham’s tube.
Glucose
fermenter but
non-lactose
fermenter.
Oxygen
requirement
Determine whether
unknown bacteria B
was aerobic or
anaerobic
Thioglycollate broth There was growth at the uppermost
part of the Thioglycollate broth.
Aerobic
Colony
morphology
Determine the
morphological
characteristics of the
unknown bacteria B
Nutrient Agar From the physical observations made
the bacteria B was rod shaped, with
colonies having chain arrangement
under microscopic examination
1mm in size,
White in
appearance,
irregular edge
shape.
3
TABLE 1: Showing different microbial tests that were performed and results obtained for
unknown bacteria B
Lab test Aim Requirements Observations made Results
Gram Stain To establish bacteria
B gram stain reaction
Crystal violet, Iodine,
Alcohol, Safranin
Purple rods were observed Gram positive
Motility test To assess if the
unknown bacteria B
was motile or non-
motile
Motility media The colonies presented with diffuse
turbid growth far from the point of
inoculation
Motile
Indole test Determine the ability
of bacteria B to
produce
tryptophanase
Sterile tryptone
water, Kovac’s
reagent
No red surface ring was formed Indole negative
Catalase Determine ability of
bacteria B to produce
catalase
Hydrogen peroxide Active bubbling was seen Catalase positive
Starch
hydrolysis
test
Determine the ability
of bacteria B to
produce amylase
Starch agar plate,
grams iodine
Clearing of light yellow/gold zone
around bacterial colonies
Amylase
production
Gelatinase
test
Determine ability of
unknown bacteria B
to produce gelatinase
Gelatin tubes, ice
water
Partial liquefaction of the inoculated
tube even after being subjected ice
water
Gelatinase
positive
Sugar
fermentatio
n
Determine the ability
of unknown bacteria
B to ferment lactose
and glucose with gas
production
Phenol Red Lactose
Broth, Phenol Red
Glucose Broth
The colour of Phenol Red Glucose
Broth changed from red to yellow
with no gas in the Durham’s tube. The
color of Phenol Red Lactose Broth
didn’t change and no gas produced in
Durham’s tube.
Glucose
fermenter but
non-lactose
fermenter.
Oxygen
requirement
Determine whether
unknown bacteria B
was aerobic or
anaerobic
Thioglycollate broth There was growth at the uppermost
part of the Thioglycollate broth.
Aerobic
Colony
morphology
Determine the
morphological
characteristics of the
unknown bacteria B
Nutrient Agar From the physical observations made
the bacteria B was rod shaped, with
colonies having chain arrangement
under microscopic examination
1mm in size,
White in
appearance,
irregular edge
shape.
3
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
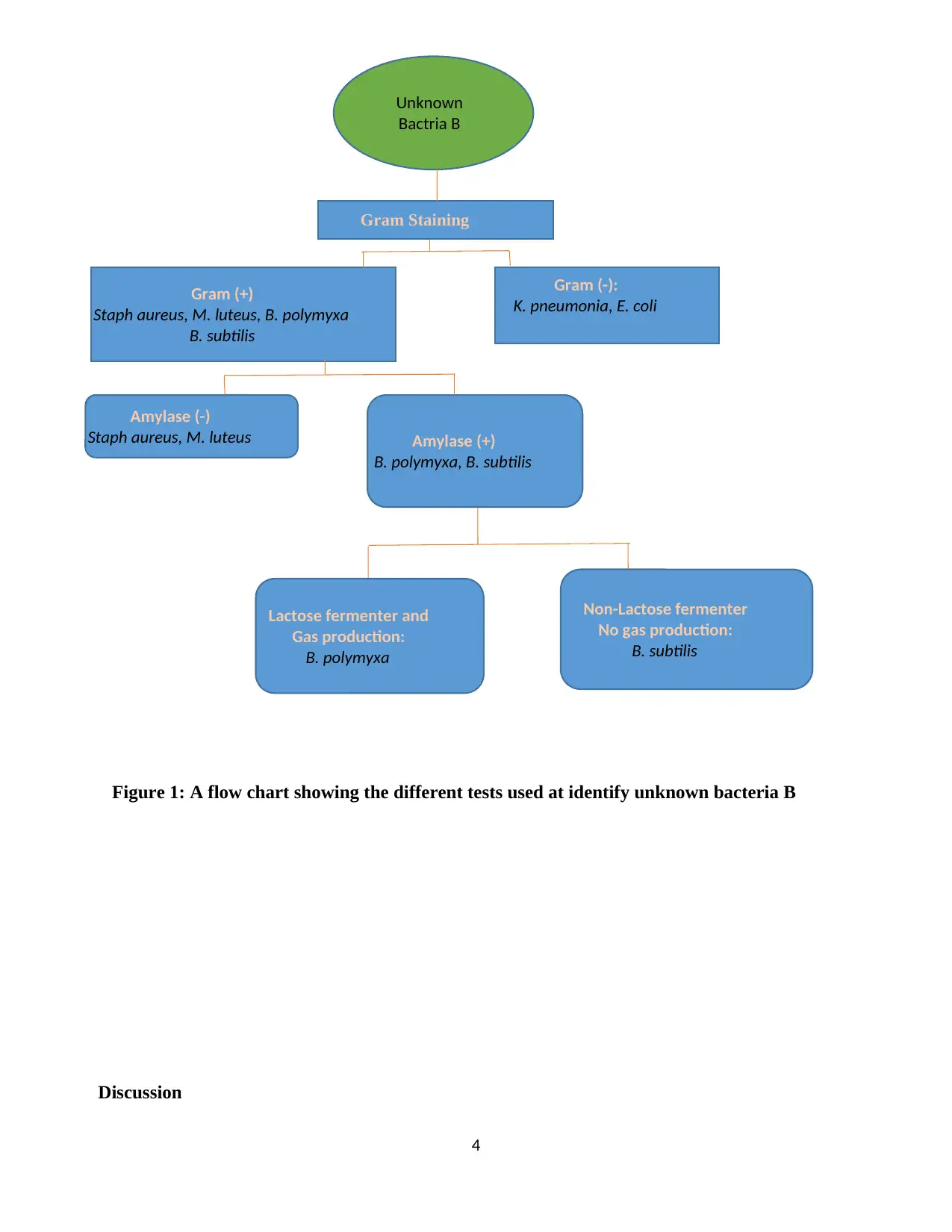
Figure 1: A flow chart showing the different tests used at identify unknown bacteria B
Discussion
4
Gram Staining
Gram (+)
Staph aureus, M. luteus, B. polymyxa
B. subtilis
Gram (-):
K. pneumonia, E. coli
Amylase (-)
Staph aureus, M. luteus Amylase (+)
B. polymyxa, B. subtilis
Lactose fermenter and
Gas production:
B. polymyxa
Non-Lactose fermenter
No gas production:
B. subtilis
Unknown
Bactria B
Discussion
4
Gram Staining
Gram (+)
Staph aureus, M. luteus, B. polymyxa
B. subtilis
Gram (-):
K. pneumonia, E. coli
Amylase (-)
Staph aureus, M. luteus Amylase (+)
B. polymyxa, B. subtilis
Lactose fermenter and
Gas production:
B. polymyxa
Non-Lactose fermenter
No gas production:
B. subtilis
Unknown
Bactria B
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Through gram staining it was possible to eliminate gram negative Enterobacteriaceae including
Escherichia coli and Klebsiella pneumonia. According to Amin, Rakhisi, and Ahmady (2015),
Bacillus subtilis are known to be present in wide range of environment including soil and human
gastrointestinal tract, they are gram positive, rod-shaped, form endospores in unfavorable
conditions. Furthermore, they are motile having a well-developed peritrichous flagella, they are
aerobic, indole negative, and catalase positive, and they are non-lactose fermenters (Khan, 2018;
Ayala et al., 2017). The other possible gram positive catalase positive bacteria was
Staphylococcus aureus. Therefore, Gelatinase and amylase test were performed on the unknown
bacteria B, the result revealed that the bacteria B was gelatinase and amylase positive further
excluding Staphylococcus aureus and Micrococcus luteus which are amylase negative (Fangio,
Roura, & Fritz, 2012). Bacillus polymyxa and Bacillus subtilis required further differential test.
Through sugar fermentation the two bacteria were differentiated, Bacillus polymyxa is both
lactose and glucose fermenter with gas production while the unknown bacteria B was not lactose
fermenter and no gas production. Therefore, this confirmed the unknown bacteria B to be Bacillus
subtilis.
Conclusion
In conclusion the unknown bacteria B was confirmed to be Bacillus subtilis based on the obtained
results. Bacillus subtilis has been extensively studied around the world. This can be attributed to
the probiotic characteristics. Moreover, Bacillus subtilis has medical importance including its
ability to produce toxic metabolites with antibiotic characteristics such as difficidin, oxydifficidin.
References
Abbott, S. L. (2011). Klebsiella, Enterobacter, Citrobacter, Serratia, Plesiomonas, and other
Enterobacteriaceae. In Manual of Clinical Microbiology, 10th Edition (pp. 639-657).
American Society of Microbiology.
Amin, M., Rakhisi, Z., & Ahmady, A. Z. (2015). Isolation and identification of Bacillus species
from soil and evaluation of their antibacterial properties. Avicenna Journal of Clinical
Microbiology and Infection, 2(1).
5
Escherichia coli and Klebsiella pneumonia. According to Amin, Rakhisi, and Ahmady (2015),
Bacillus subtilis are known to be present in wide range of environment including soil and human
gastrointestinal tract, they are gram positive, rod-shaped, form endospores in unfavorable
conditions. Furthermore, they are motile having a well-developed peritrichous flagella, they are
aerobic, indole negative, and catalase positive, and they are non-lactose fermenters (Khan, 2018;
Ayala et al., 2017). The other possible gram positive catalase positive bacteria was
Staphylococcus aureus. Therefore, Gelatinase and amylase test were performed on the unknown
bacteria B, the result revealed that the bacteria B was gelatinase and amylase positive further
excluding Staphylococcus aureus and Micrococcus luteus which are amylase negative (Fangio,
Roura, & Fritz, 2012). Bacillus polymyxa and Bacillus subtilis required further differential test.
Through sugar fermentation the two bacteria were differentiated, Bacillus polymyxa is both
lactose and glucose fermenter with gas production while the unknown bacteria B was not lactose
fermenter and no gas production. Therefore, this confirmed the unknown bacteria B to be Bacillus
subtilis.
Conclusion
In conclusion the unknown bacteria B was confirmed to be Bacillus subtilis based on the obtained
results. Bacillus subtilis has been extensively studied around the world. This can be attributed to
the probiotic characteristics. Moreover, Bacillus subtilis has medical importance including its
ability to produce toxic metabolites with antibiotic characteristics such as difficidin, oxydifficidin.
References
Abbott, S. L. (2011). Klebsiella, Enterobacter, Citrobacter, Serratia, Plesiomonas, and other
Enterobacteriaceae. In Manual of Clinical Microbiology, 10th Edition (pp. 639-657).
American Society of Microbiology.
Amin, M., Rakhisi, Z., & Ahmady, A. Z. (2015). Isolation and identification of Bacillus species
from soil and evaluation of their antibacterial properties. Avicenna Journal of Clinical
Microbiology and Infection, 2(1).
5

Ayala, F. R., Bauman, C., Cogliati, S., Leñini, C., Bartolini, M., & Grau, R. (2017). Microbial
flora, probiotics, Bacillus subtilis and the search for a long and healthy human longevity.
Microbial Cell, 4(4), 133.
Cheesbrough, M. (2006). District laboratory practice in tropical countries. Cambridge university
press.
Cowan, S. T. (2004). Cowan and Steel's manual for the identification of medical bacteria.
Cambridge university press.
Fangio, M. F., Roura, S. I., & Fritz, R. (2010). Isolation and identification of Bacillus spp. and
related genera from different starchy foods. Journal of food science, 75(4), M218-M221.
Khan, K. (2018). Isolation of bacteria from dairy-based popular sweetmeat named swandesh
following the molecular identification of Bacillus species (Doctoral dissertation, BRAC
Univeristy).
Luna, V. A., Peak, K. K., Veguilla, W. O., Reeves, F., Heberlein-Larson, L., Cannons, A. C.
Cattani, J. (2005). Use of Two Selective Media and a Broth Motility Test Can Aid in
Identification or Exclusion of Bacillus anthracis. Journal of Clinical Microbiology, 43(9),
4336–4341. http://doi.org/10.1128/JCM.43.9.4336-4341.2005
Pogmore, A. R., Seistrup, K. H., & Strahl, H. (2018). The Gram-positive model organism Bacillus
subtilis does not form microscopically detectable cardiolipin-specific lipid domains.
bioRxiv, 190306.
Xia, Y., Kong, Y., Seviour, R., Yang, H. E., Forster, R., Vasanthan, T., & McAllister, T. (2015). In
situ identification and quantification of starch-hydrolyzing bacteria attached to barley and
corn grain in the rumen of cows fed barley-based diets. FEMS microbiology ecology,
91(8).
6
flora, probiotics, Bacillus subtilis and the search for a long and healthy human longevity.
Microbial Cell, 4(4), 133.
Cheesbrough, M. (2006). District laboratory practice in tropical countries. Cambridge university
press.
Cowan, S. T. (2004). Cowan and Steel's manual for the identification of medical bacteria.
Cambridge university press.
Fangio, M. F., Roura, S. I., & Fritz, R. (2010). Isolation and identification of Bacillus spp. and
related genera from different starchy foods. Journal of food science, 75(4), M218-M221.
Khan, K. (2018). Isolation of bacteria from dairy-based popular sweetmeat named swandesh
following the molecular identification of Bacillus species (Doctoral dissertation, BRAC
Univeristy).
Luna, V. A., Peak, K. K., Veguilla, W. O., Reeves, F., Heberlein-Larson, L., Cannons, A. C.
Cattani, J. (2005). Use of Two Selective Media and a Broth Motility Test Can Aid in
Identification or Exclusion of Bacillus anthracis. Journal of Clinical Microbiology, 43(9),
4336–4341. http://doi.org/10.1128/JCM.43.9.4336-4341.2005
Pogmore, A. R., Seistrup, K. H., & Strahl, H. (2018). The Gram-positive model organism Bacillus
subtilis does not form microscopically detectable cardiolipin-specific lipid domains.
bioRxiv, 190306.
Xia, Y., Kong, Y., Seviour, R., Yang, H. E., Forster, R., Vasanthan, T., & McAllister, T. (2015). In
situ identification and quantification of starch-hydrolyzing bacteria attached to barley and
corn grain in the rumen of cows fed barley-based diets. FEMS microbiology ecology,
91(8).
6
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 6
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.