Lab Report on Green Fluorescent Protein (GFP)
VerifiedAdded on 2023/04/20
|19
|3618
|186
AI Summary
This lab report focuses on the analysis and study of green fluorescent protein (GFPuv gene) using fluorimetry and mass spectroscopy. Site directed mutagenesis was performed to alter the functions and properties of GFP. The report discusses the cloning and sub-cloning of the GFPuv gene, transformation of ligated products into E. coli, and the induction and purification of GFP protein.
Contribute Materials
Your contribution can guide someone’s learning journey. Share your
documents today.

Running head: LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
Production of different spectral characteristics of green fluorescent protein using gene
engineering
Name of the Student
Name of the University
Author’s Note:
Production of different spectral characteristics of green fluorescent protein using gene
engineering
Name of the Student
Name of the University
Author’s Note:
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

1LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
Abstract
Green fluorescent protein is present among the jellyfish (Aequorea Victoria) found in the
north pacific and it is a readily available protein and can be used to track the gene expression
owing to the structure and stability of this protein’s choromophore. The aim of this
investigation is to analyse and study the properties of green fluorescent protein (GFPuv gene)
by fluorimetry and mass spectroscopy. Additionally, site directed mutagenesis was performed
to further alter the functions and properties of green fluorescent protein. In this study, two
GFP mutations were achieved which were able to generate the EGFP and the size of the
EGFP was determined at 29 kDa. Further study of EGFP with different cell types might be
useful for better understanding of the properties of EGFP.
Keywords: GFP, Site Direct Mutagenesis, EGFP, Mass Spectrophotometer and
Fluorometry.
Abstract
Green fluorescent protein is present among the jellyfish (Aequorea Victoria) found in the
north pacific and it is a readily available protein and can be used to track the gene expression
owing to the structure and stability of this protein’s choromophore. The aim of this
investigation is to analyse and study the properties of green fluorescent protein (GFPuv gene)
by fluorimetry and mass spectroscopy. Additionally, site directed mutagenesis was performed
to further alter the functions and properties of green fluorescent protein. In this study, two
GFP mutations were achieved which were able to generate the EGFP and the size of the
EGFP was determined at 29 kDa. Further study of EGFP with different cell types might be
useful for better understanding of the properties of EGFP.
Keywords: GFP, Site Direct Mutagenesis, EGFP, Mass Spectrophotometer and
Fluorometry.

2LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
Table of Contents
Introduction:...........................................................................................................................3
Materials and Methods:..........................................................................................................5
Cloning and Sub- cloning of GFPuv gene using expression vector:.................................5
Transformation of ligated products into DH5α cells:........................................................5
Site directed mutagenesis:..................................................................................................5
Induction of GFP protein expression:................................................................................6
His-tagged EGFP purification by immobilized metal affinity chromatography:...............7
EGFP characterisation by Fluorimetry and Mass Spectrometry:.......................................7
Results:...................................................................................................................................8
Cloning and Sub- cloning of GFPuv gene and transformation of ligated products in to E.
coli:.....................................................................................................................................8
Site Directed mutagenesis:.................................................................................................8
Mutant protein expression:...............................................................................................10
Protein purification:.........................................................................................................12
Protein Analysis:..............................................................................................................13
Discussion:...........................................................................................................................13
References:...........................................................................................................................16
Table of Contents
Introduction:...........................................................................................................................3
Materials and Methods:..........................................................................................................5
Cloning and Sub- cloning of GFPuv gene using expression vector:.................................5
Transformation of ligated products into DH5α cells:........................................................5
Site directed mutagenesis:..................................................................................................5
Induction of GFP protein expression:................................................................................6
His-tagged EGFP purification by immobilized metal affinity chromatography:...............7
EGFP characterisation by Fluorimetry and Mass Spectrometry:.......................................7
Results:...................................................................................................................................8
Cloning and Sub- cloning of GFPuv gene and transformation of ligated products in to E.
coli:.....................................................................................................................................8
Site Directed mutagenesis:.................................................................................................8
Mutant protein expression:...............................................................................................10
Protein purification:.........................................................................................................12
Protein Analysis:..............................................................................................................13
Discussion:...........................................................................................................................13
References:...........................................................................................................................16

3LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
Introduction:
GFP or green fluorescent protein is kind of bioluminescent polypeptide which is present
among the jellyfish (Aequorea Victoria) found in the north pacific. This protein converts the
blue light emitted by a bioluminescent protein present in the jellyfish in to green light.
Purified solution of this protein looks yellow but under direct sunlight it produces a vibrant
green colour by absorbing ultraviolet ray present in the sunlight. This protein was first
discovered in the year 1962 and the gene that encodes this green fluorescent protein was first
cloned in the year 1990 (Prasher et al., 1992). Green fluorescent protein is comprises of 238
residues and weighs 21 kDa. The structure of this green fluorescent protein contains one
eleven stranded β- pleated sheet and five α- helices (Zimmer, 2002). The 3D structure of this
green fluorescent protein can be seen in the Figure 1 below.
Figure 1: The 3D structure of the green fluorescent protein (Source: RCSB Bank, 2019)
Introduction:
GFP or green fluorescent protein is kind of bioluminescent polypeptide which is present
among the jellyfish (Aequorea Victoria) found in the north pacific. This protein converts the
blue light emitted by a bioluminescent protein present in the jellyfish in to green light.
Purified solution of this protein looks yellow but under direct sunlight it produces a vibrant
green colour by absorbing ultraviolet ray present in the sunlight. This protein was first
discovered in the year 1962 and the gene that encodes this green fluorescent protein was first
cloned in the year 1990 (Prasher et al., 1992). Green fluorescent protein is comprises of 238
residues and weighs 21 kDa. The structure of this green fluorescent protein contains one
eleven stranded β- pleated sheet and five α- helices (Zimmer, 2002). The 3D structure of this
green fluorescent protein can be seen in the Figure 1 below.
Figure 1: The 3D structure of the green fluorescent protein (Source: RCSB Bank, 2019)
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

4LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
Each stand of this β- pleated sheet contains 9 to 13 residues. Through hydrogen bonding, this
β- pleated sheet conforms into β- barrel. Two α- helices are present at the both end of this
molecule which helps form the structure in to a perfect cylinder. One α- helix run through the
central axis of the β- barrel approximately in 90 degree angle and this strand of the α- helix is
responsible for fluorescence as it contains the fluorophore. This fluorophore is generally
comprised of three amino acids namely serine- tyrosine- glycine. However, sometimes, the
amino acid serine in this structure is replaced with threonine.In case of the wild type, the
chromophore for this green fluorescent protein has a peak excitation at 395 nm and a minor
excitation peak at 475 nm. The intensity is around 3 times less in case of the minor excitation
peak (Zimmer, 2002).
Green fluorescent protein is a readily available protein and can be used to track the gene
expression owing to the structure and stability of this protein’s choromophore. Along with
that, this gene does not interfere with the function of living organism during its expression in
them (Cody et al., 1993). Hence, it is extremely useful to the scientists and researchers
investigating in the area of living cells. Scientists can now engineer yellow and blue
fluorescent protein using this protein. This can have usefulness in the medical research as
well. This green fluorescent protein can be used to study and monitor the progressions of
diseases like Leishmania (Lang et al., 2005). Additionally, scientists are using this protein to
create biosensors.
Although, this green fluorescent protein have some limitations as well which can hinder the
advantages of the green fluorescent protein. For example, this protein folding efficacy
decreases at higher temperature (Thomas & Maule, 2000). However, the advantages are far
outweighs the disadvantages present in this protein.
Each stand of this β- pleated sheet contains 9 to 13 residues. Through hydrogen bonding, this
β- pleated sheet conforms into β- barrel. Two α- helices are present at the both end of this
molecule which helps form the structure in to a perfect cylinder. One α- helix run through the
central axis of the β- barrel approximately in 90 degree angle and this strand of the α- helix is
responsible for fluorescence as it contains the fluorophore. This fluorophore is generally
comprised of three amino acids namely serine- tyrosine- glycine. However, sometimes, the
amino acid serine in this structure is replaced with threonine.In case of the wild type, the
chromophore for this green fluorescent protein has a peak excitation at 395 nm and a minor
excitation peak at 475 nm. The intensity is around 3 times less in case of the minor excitation
peak (Zimmer, 2002).
Green fluorescent protein is a readily available protein and can be used to track the gene
expression owing to the structure and stability of this protein’s choromophore. Along with
that, this gene does not interfere with the function of living organism during its expression in
them (Cody et al., 1993). Hence, it is extremely useful to the scientists and researchers
investigating in the area of living cells. Scientists can now engineer yellow and blue
fluorescent protein using this protein. This can have usefulness in the medical research as
well. This green fluorescent protein can be used to study and monitor the progressions of
diseases like Leishmania (Lang et al., 2005). Additionally, scientists are using this protein to
create biosensors.
Although, this green fluorescent protein have some limitations as well which can hinder the
advantages of the green fluorescent protein. For example, this protein folding efficacy
decreases at higher temperature (Thomas & Maule, 2000). However, the advantages are far
outweighs the disadvantages present in this protein.

5LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
Therefore, the aim of this investigation is to analyse and study the properties of green
fluorescent protein (GFPuv gene) by fluorimetry and mass spectroscopy. Additionally, site
directed mutagenesis was performed to further alter the functions and properties of green
fluorescent protein.
Materials and Methods:
Cloning and Sub- cloning of GFPuv gene using expression vector:
To clone the GFP gene pET23 plasmid was used and the size of the plasmids was 3592 bp.
Open reading GFPuv frame was inserted between the restriction sites of the pET23 to
construct the pET23gfpuv. These two restriction sites are Hindll and EcoRI. At first, an Ndel
restriction site was inserted by the addition of 15 ml of DNA plasmid. After that, the products
were separated using 1.2 per cent agarose gel and were analysed. 761 bp long GFPuv
fragment were identified using a UV transilluminator and was extracted by using the
QIAquick gel extraction kit (Qiagen). The sub clone of the GFPuv gene, pET28c expression
vector was used. In order to achieve this, GFPuv fragment and pET28c (Novagen) was
digested together in order to have a compatible cohesive end which is needed for the
formation of circular recombinant DNA. T4 DNA ligase was used for the ligation of GFPuv
and pET28c.
Transformation of ligated products into DH5α cells:
After the completion of ligation reaction, DHα cells of E. coli were used for the
transformation of the ligation product. The replication of these product
was conducted by heat shocking. The heat shocking step was performed
at 420C and time duration was for 90 seconds. After that, the cells were
incubated at 37 0 C overnight and grown in the SOS media. LB plates were
Therefore, the aim of this investigation is to analyse and study the properties of green
fluorescent protein (GFPuv gene) by fluorimetry and mass spectroscopy. Additionally, site
directed mutagenesis was performed to further alter the functions and properties of green
fluorescent protein.
Materials and Methods:
Cloning and Sub- cloning of GFPuv gene using expression vector:
To clone the GFP gene pET23 plasmid was used and the size of the plasmids was 3592 bp.
Open reading GFPuv frame was inserted between the restriction sites of the pET23 to
construct the pET23gfpuv. These two restriction sites are Hindll and EcoRI. At first, an Ndel
restriction site was inserted by the addition of 15 ml of DNA plasmid. After that, the products
were separated using 1.2 per cent agarose gel and were analysed. 761 bp long GFPuv
fragment were identified using a UV transilluminator and was extracted by using the
QIAquick gel extraction kit (Qiagen). The sub clone of the GFPuv gene, pET28c expression
vector was used. In order to achieve this, GFPuv fragment and pET28c (Novagen) was
digested together in order to have a compatible cohesive end which is needed for the
formation of circular recombinant DNA. T4 DNA ligase was used for the ligation of GFPuv
and pET28c.
Transformation of ligated products into DH5α cells:
After the completion of ligation reaction, DHα cells of E. coli were used for the
transformation of the ligation product. The replication of these product
was conducted by heat shocking. The heat shocking step was performed
at 420C and time duration was for 90 seconds. After that, the cells were
incubated at 37 0 C overnight and grown in the SOS media. LB plates were

6LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
used for this purpose and plates contained the kanamycin antibiotic for
the selection of pET28c plasmid containing colonies.
Site directed mutagenesis:
Quick change site directed mutagenesis method was adopted for the enhancement of the
green fluorescent protein (GFP). This is known as enhanced GFP or EGFP. Stratagene
developed procedure was adopted for this purpose the 64 and 65 position in amino acid were
targeted for the generation of enhanced GFP (EGFP). Merck supplied KOD DNA
polymerase was used in this procedure. For the PCR procedure, following
materials were used: 10x PCR buffer, 2millimeter dNTP mix, 20ng of
GFPuv -Pet28c templet DNA, 1U hot start KOD polymerase, 33millimeter
sterile water, 25millimeter MgSO4, and 125ng each of the Reverse prime
and Forward primer. The sequences of the Reverse prime and forward
primer are as follows:
M2R
5’-CGGGAAAAGCATTGAACACCATAAGTGAGAGTAGTGACAAGTG-3
M2F
5’-CACTTGTCACTACTCTCACTTATGGTGTTCAATGCTTTTCCCG-3’
After the completion of the PCR, the resultant products were digested and incubated for 1
hour at 37 0 C. The digestion was performed using Dpn1 (10U). XI-1 blue super-
competent cells were used for the transformation of the Dpn1 digest
products. After that, the DH5α bacterial cells were used for the
transformation of the mutant recombinant vector. This procedure was
performed using standard protocol.
used for this purpose and plates contained the kanamycin antibiotic for
the selection of pET28c plasmid containing colonies.
Site directed mutagenesis:
Quick change site directed mutagenesis method was adopted for the enhancement of the
green fluorescent protein (GFP). This is known as enhanced GFP or EGFP. Stratagene
developed procedure was adopted for this purpose the 64 and 65 position in amino acid were
targeted for the generation of enhanced GFP (EGFP). Merck supplied KOD DNA
polymerase was used in this procedure. For the PCR procedure, following
materials were used: 10x PCR buffer, 2millimeter dNTP mix, 20ng of
GFPuv -Pet28c templet DNA, 1U hot start KOD polymerase, 33millimeter
sterile water, 25millimeter MgSO4, and 125ng each of the Reverse prime
and Forward primer. The sequences of the Reverse prime and forward
primer are as follows:
M2R
5’-CGGGAAAAGCATTGAACACCATAAGTGAGAGTAGTGACAAGTG-3
M2F
5’-CACTTGTCACTACTCTCACTTATGGTGTTCAATGCTTTTCCCG-3’
After the completion of the PCR, the resultant products were digested and incubated for 1
hour at 37 0 C. The digestion was performed using Dpn1 (10U). XI-1 blue super-
competent cells were used for the transformation of the Dpn1 digest
products. After that, the DH5α bacterial cells were used for the
transformation of the mutant recombinant vector. This procedure was
performed using standard protocol.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

7LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
Induction of GFP protein expression:
To induce the expression of the GFP protein, a glucose and lactose mixed media (SB-
5052) was used. The media contained 100 μg/ ml of kanamycin antibiotic and 15
μl of the culture was added to 25ml SM-5052. After that the culture was
incubated for 20 hours at 37 0 C while shaking at 350 – 400 rpm. Induced
and non- induced samples were re- suspended and harvested in 100μl of
SDS- PAGE buffer. Novagen make BUGBUSTER Protein Extraction Reagent
was used for the fractionation of the culture into insoluble and soluble
part. Then, all these samples were run on a polyacrylamide gel. The
running condition was for 1 hour at 150 V. Western blotting was
performed on the fractionated samples to detect the expression of
enhanced green fluorescence protein. Novagen make HisprobeA®-HRP
was used for the probing the western blot. Chemiluminescent kit was used
to analyze the western blot after washing with TBST.
His-tagged EGFP purification by immobilized metal affinity chromatography:
Affinity chromatography (Ni- NTA) was used for the purification purpose of the fractionated
soluble parts of the His-tagged GFP protein. Agarose resin packed column was
used to load the samples and this resin was used for the removal of the
non- specific protein binding by washing with deionised water. After the
purification and elution, the protein concentration of the elution part was
measured using Bradford method and for the measurement of the protein
purity, SDS- PAGE procedure was applied.
Induction of GFP protein expression:
To induce the expression of the GFP protein, a glucose and lactose mixed media (SB-
5052) was used. The media contained 100 μg/ ml of kanamycin antibiotic and 15
μl of the culture was added to 25ml SM-5052. After that the culture was
incubated for 20 hours at 37 0 C while shaking at 350 – 400 rpm. Induced
and non- induced samples were re- suspended and harvested in 100μl of
SDS- PAGE buffer. Novagen make BUGBUSTER Protein Extraction Reagent
was used for the fractionation of the culture into insoluble and soluble
part. Then, all these samples were run on a polyacrylamide gel. The
running condition was for 1 hour at 150 V. Western blotting was
performed on the fractionated samples to detect the expression of
enhanced green fluorescence protein. Novagen make HisprobeA®-HRP
was used for the probing the western blot. Chemiluminescent kit was used
to analyze the western blot after washing with TBST.
His-tagged EGFP purification by immobilized metal affinity chromatography:
Affinity chromatography (Ni- NTA) was used for the purification purpose of the fractionated
soluble parts of the His-tagged GFP protein. Agarose resin packed column was
used to load the samples and this resin was used for the removal of the
non- specific protein binding by washing with deionised water. After the
purification and elution, the protein concentration of the elution part was
measured using Bradford method and for the measurement of the protein
purity, SDS- PAGE procedure was applied.

8LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
EGFP characterisation by Fluorimetry and Mass Spectrometry:
Properties of EGFP were analysed using mass spectrophometry. In order to do that, 3.5 KV
ESI capillary was used as a drying gas and the nebulising gas was nitrogen. 30μL of the
protein sample was treated with 1 per cent aqueous formic acid and 15 μL
methanol was used as sample for the mass spectrophotometry. Then,
ratio of mass to charge was calculated from the collected data.
For the fluorometric analysis, following parameters were used:
Emission wavelength between 490 and 550 nm.
Excitation wavelength at 450 nm.
Slit width – 2.5 nm
Scan rate – 1 nm/ sec
Accumulation rate – 3
Results:
Cloning and Sub- cloning of GFPuv gene and transformation of ligated products in to
E. coli:
GFPuv sub cloning was performed successfully and UV spectrophotometer was used for the
measurement of the pET23-gfpuv DNA concentration. The purity ratio of the
pET23-gfpuv DNA was 2 and the concentration was at 1.8ng. The ligated
products which were transformed into E. coli cells had appeared as white
colonies on the LB- kanamycin plates. This signifies the presence of the recombined
plasmids of pET28c-gfpuv containing colonies on the LB- kanamycin plates as
well as the control plate containing the undigested pET28c plasmid. The efficacy of
this transformation was 10.6 per cent.
EGFP characterisation by Fluorimetry and Mass Spectrometry:
Properties of EGFP were analysed using mass spectrophometry. In order to do that, 3.5 KV
ESI capillary was used as a drying gas and the nebulising gas was nitrogen. 30μL of the
protein sample was treated with 1 per cent aqueous formic acid and 15 μL
methanol was used as sample for the mass spectrophotometry. Then,
ratio of mass to charge was calculated from the collected data.
For the fluorometric analysis, following parameters were used:
Emission wavelength between 490 and 550 nm.
Excitation wavelength at 450 nm.
Slit width – 2.5 nm
Scan rate – 1 nm/ sec
Accumulation rate – 3
Results:
Cloning and Sub- cloning of GFPuv gene and transformation of ligated products in to
E. coli:
GFPuv sub cloning was performed successfully and UV spectrophotometer was used for the
measurement of the pET23-gfpuv DNA concentration. The purity ratio of the
pET23-gfpuv DNA was 2 and the concentration was at 1.8ng. The ligated
products which were transformed into E. coli cells had appeared as white
colonies on the LB- kanamycin plates. This signifies the presence of the recombined
plasmids of pET28c-gfpuv containing colonies on the LB- kanamycin plates as
well as the control plate containing the undigested pET28c plasmid. The efficacy of
this transformation was 10.6 per cent.

9LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
Site Directed mutagenesis:
Qiagen make QlA prep Kit was used for the generation of the EGFP. Positive
was observed on the culture plates. The sequencing result approves the
presence of mutation in Ser65Thr and Phe64Leu gene. The result can be
seen in the Figure 1 and Figure 2.
From the Figure 2, it can be seen that the amino acid sequence for GFPuv
was aligned with the wild type. Amino acid F is mutated to amino acid L at
the position 66. Additionally, the amino acid S is mutated to the amino
acid T at the position 67. Therefore, it can be stated that the EGFP primers
were mutated and would work in this sequence.
Site Directed mutagenesis:
Qiagen make QlA prep Kit was used for the generation of the EGFP. Positive
was observed on the culture plates. The sequencing result approves the
presence of mutation in Ser65Thr and Phe64Leu gene. The result can be
seen in the Figure 1 and Figure 2.
From the Figure 2, it can be seen that the amino acid sequence for GFPuv
was aligned with the wild type. Amino acid F is mutated to amino acid L at
the position 66. Additionally, the amino acid S is mutated to the amino
acid T at the position 67. Therefore, it can be stated that the EGFP primers
were mutated and would work in this sequence.
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

10LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
Figure 1: Site- directed Mutagenesis
Figure 1: Site- directed Mutagenesis

11LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
Figure 2: Site directed mutagenesis (Continuation from the Figure 1)
Mutant protein expression:
Presence of EGFP was verified by the soluble fraction SDS- PAGE. From the results, it can
be verified the presence of putative EGFP and the size of the EGFP was at around 27 kDa.
The SDS- PAGE result obtained from this experiment can be seen in the Figure 3. Presence
of putative EGFP can be seen in the well three and fourth. Induced samples were provided in
Figure 2: Site directed mutagenesis (Continuation from the Figure 1)
Mutant protein expression:
Presence of EGFP was verified by the soluble fraction SDS- PAGE. From the results, it can
be verified the presence of putative EGFP and the size of the EGFP was at around 27 kDa.
The SDS- PAGE result obtained from this experiment can be seen in the Figure 3. Presence
of putative EGFP can be seen in the well three and fourth. Induced samples were provided in
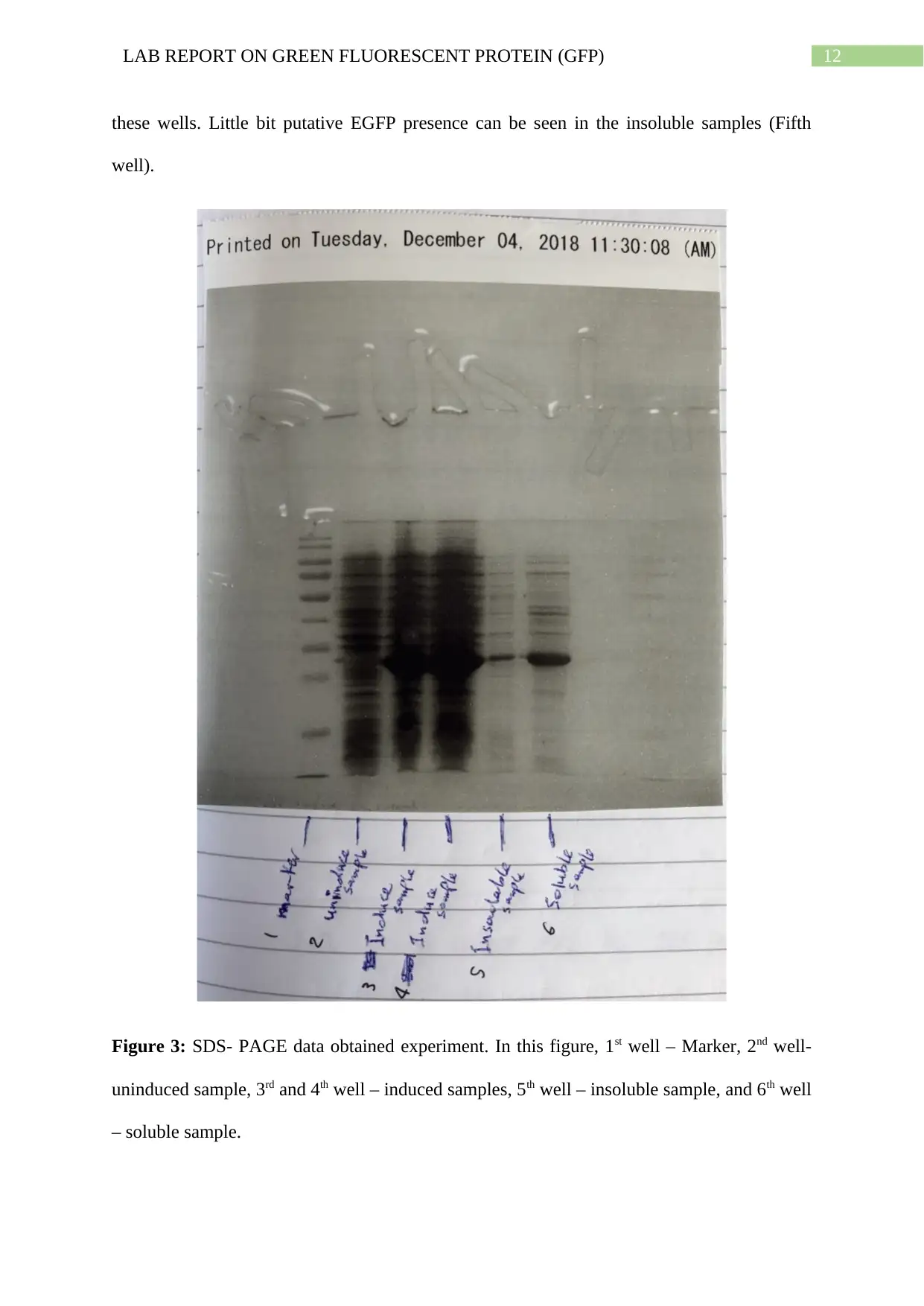
12LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
these wells. Little bit putative EGFP presence can be seen in the insoluble samples (Fifth
well).
Figure 3: SDS- PAGE data obtained experiment. In this figure, 1st well – Marker, 2nd well-
uninduced sample, 3rd and 4th well – induced samples, 5th well – insoluble sample, and 6th well
– soluble sample.
these wells. Little bit putative EGFP presence can be seen in the insoluble samples (Fifth
well).
Figure 3: SDS- PAGE data obtained experiment. In this figure, 1st well – Marker, 2nd well-
uninduced sample, 3rd and 4th well – induced samples, 5th well – insoluble sample, and 6th well
– soluble sample.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

13LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
Finally, a thick band can be seen in case of soluble sample (Sixth well) owing to the reason of
overexpression. These data obtained from the SDS- PAGE follows the same trend of data
collected from Western Blot. Western Blot results can be seen in the Figure 4.
.
Figure 4: Presentation of data collected from Western Blot.
Protein purification:
Bradford method was used to determine the concentration of His Tag-EGFP protein and
it is evident from the result that the residue of 6 histidine amino acids is
presence in the expression of pET28c vector. The concentration of in the
elution part of the His Tag-EGFP protein was around 2.12 mg/ ml and
purity was confirmed by SDS-PAGE. The appearance of majority of EGFP is
Finally, a thick band can be seen in case of soluble sample (Sixth well) owing to the reason of
overexpression. These data obtained from the SDS- PAGE follows the same trend of data
collected from Western Blot. Western Blot results can be seen in the Figure 4.
.
Figure 4: Presentation of data collected from Western Blot.
Protein purification:
Bradford method was used to determine the concentration of His Tag-EGFP protein and
it is evident from the result that the residue of 6 histidine amino acids is
presence in the expression of pET28c vector. The concentration of in the
elution part of the His Tag-EGFP protein was around 2.12 mg/ ml and
purity was confirmed by SDS-PAGE. The appearance of majority of EGFP is
14LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
expected in the first elution as it is the first elution and correspondingly,
small amount of protein has been observed after the second elution.
Protein Analysis:
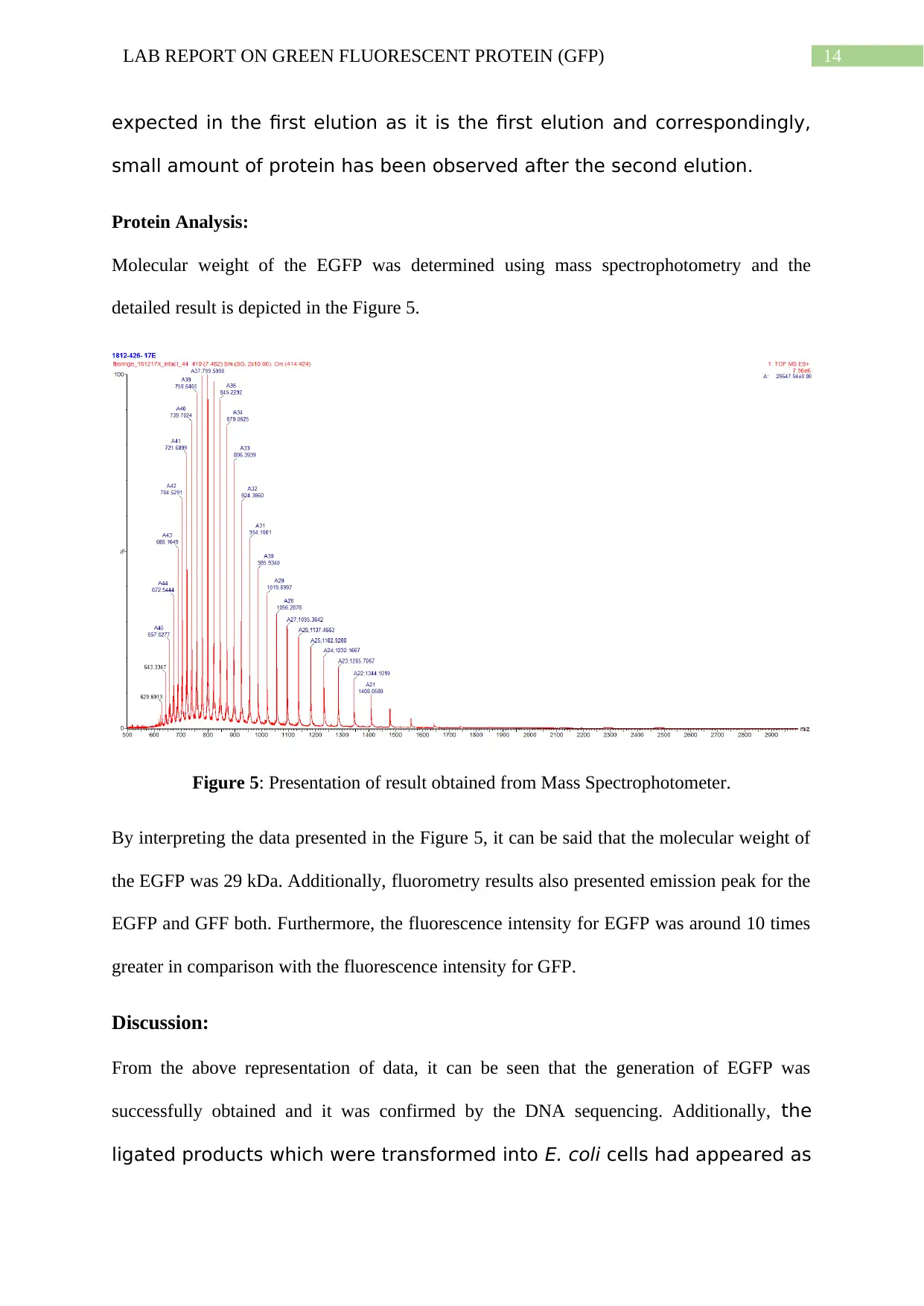
Molecular weight of the EGFP was determined using mass spectrophotometry and the
detailed result is depicted in the Figure 5.
Figure 5: Presentation of result obtained from Mass Spectrophotometer.
By interpreting the data presented in the Figure 5, it can be said that the molecular weight of
the EGFP was 29 kDa. Additionally, fluorometry results also presented emission peak for the
EGFP and GFF both. Furthermore, the fluorescence intensity for EGFP was around 10 times
greater in comparison with the fluorescence intensity for GFP.
Discussion:
From the above representation of data, it can be seen that the generation of EGFP was
successfully obtained and it was confirmed by the DNA sequencing. Additionally, the
ligated products which were transformed into E. coli cells had appeared as
expected in the first elution as it is the first elution and correspondingly,
small amount of protein has been observed after the second elution.
Protein Analysis:
Molecular weight of the EGFP was determined using mass spectrophotometry and the
detailed result is depicted in the Figure 5.
Figure 5: Presentation of result obtained from Mass Spectrophotometer.
By interpreting the data presented in the Figure 5, it can be said that the molecular weight of
the EGFP was 29 kDa. Additionally, fluorometry results also presented emission peak for the
EGFP and GFF both. Furthermore, the fluorescence intensity for EGFP was around 10 times
greater in comparison with the fluorescence intensity for GFP.
Discussion:
From the above representation of data, it can be seen that the generation of EGFP was
successfully obtained and it was confirmed by the DNA sequencing. Additionally, the
ligated products which were transformed into E. coli cells had appeared as

15LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
white colonies on the LB- kanamycin plates. This signifies the presence of the
recombined pET28c-gfpuv plasmids in those colonies. Therefore, it can be
inferred that the auto induction was successful in case of this
investigation. The presence of the EGFP band for the induced sample in
western blot also validates this suggestion. SDS- PAGE results also
confirm the purification of protein obtained from this experiment. In this
investigation, it has been found that the fluorescence intensity for EGFP was
around 10 times greater in comparison with the fluorescence intensity for GFP. This is
similar to the findings published in this research area (Cormack et al., 1996). Studies
have reported that the fluorescence intensity for EGFP is generally higher than the
fluorescence intensity for GFP (Tsien, 1998). However, in this investigation the difference is
at around 10 times for EGFP in comparison with the GFP. This occurrence can be explained
by the fact that mutation EGFP ionized the chromospore and thus, it is providing higher
brightness.
The fluorometry result obtained in this study also is in line with the findings presented by the
other investigators who have been investigating in this area. The emission peak for GFP was
observed at 504 nm and the emission peak for the EGFP was observed at the 507 nm. It has
been reported in the published journal that the emission peak of the wild type GFP was at 504
nm and emission peak of mutated EGFP was in the range of 507 to 509 nm while the folding
efficiency was at 20 (Patterson, Day & Piston, 2001).
In the published study, the molecular weight of the EGFP has been mentioned as 27 kDa
(Tsien, 1998). This is also in line with the findings from this investigation and molecular
weight of EGFP has been calculated from this investigation was 29 kDa. The extra weight of
the EGFP might happened due to the addition of six additional histidine tag and methalonin.
white colonies on the LB- kanamycin plates. This signifies the presence of the
recombined pET28c-gfpuv plasmids in those colonies. Therefore, it can be
inferred that the auto induction was successful in case of this
investigation. The presence of the EGFP band for the induced sample in
western blot also validates this suggestion. SDS- PAGE results also
confirm the purification of protein obtained from this experiment. In this
investigation, it has been found that the fluorescence intensity for EGFP was
around 10 times greater in comparison with the fluorescence intensity for GFP. This is
similar to the findings published in this research area (Cormack et al., 1996). Studies
have reported that the fluorescence intensity for EGFP is generally higher than the
fluorescence intensity for GFP (Tsien, 1998). However, in this investigation the difference is
at around 10 times for EGFP in comparison with the GFP. This occurrence can be explained
by the fact that mutation EGFP ionized the chromospore and thus, it is providing higher
brightness.
The fluorometry result obtained in this study also is in line with the findings presented by the
other investigators who have been investigating in this area. The emission peak for GFP was
observed at 504 nm and the emission peak for the EGFP was observed at the 507 nm. It has
been reported in the published journal that the emission peak of the wild type GFP was at 504
nm and emission peak of mutated EGFP was in the range of 507 to 509 nm while the folding
efficiency was at 20 (Patterson, Day & Piston, 2001).
In the published study, the molecular weight of the EGFP has been mentioned as 27 kDa
(Tsien, 1998). This is also in line with the findings from this investigation and molecular
weight of EGFP has been calculated from this investigation was 29 kDa. The extra weight of
the EGFP might happened due to the addition of six additional histidine tag and methalonin.
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

16LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
In this investigation, treble mutation of GFPuv has been achieved and this can be used for the
improvement of the research in the area of cell biology. New variant of GFP mutations can
have significant advantages if the spectral properties are properly studied (Chalfie et al.,
1994). Therefore, study of EGFP with different cell types might be useful for better
understanding of the properties of EGFP. The strength of this study is that it has reported two
GFP mutations which were able to generate the EGFP.
Therefore, from the above discussion, it can be concluded that this study has fulfilled its aim
which was to generate EGFP through site directed mutagenesis. Additionally, two GFP
mutations were achieved which were able to generate the EGFP. Furthermore, the findings
correlate the findings published by other authors in this research area. Therefore, the findings
obtained from this study can be used by future investigators investigating in this area and
study of EGFP with different cell types might be useful for better understanding of the
properties of EGFP.
In this investigation, treble mutation of GFPuv has been achieved and this can be used for the
improvement of the research in the area of cell biology. New variant of GFP mutations can
have significant advantages if the spectral properties are properly studied (Chalfie et al.,
1994). Therefore, study of EGFP with different cell types might be useful for better
understanding of the properties of EGFP. The strength of this study is that it has reported two
GFP mutations which were able to generate the EGFP.
Therefore, from the above discussion, it can be concluded that this study has fulfilled its aim
which was to generate EGFP through site directed mutagenesis. Additionally, two GFP
mutations were achieved which were able to generate the EGFP. Furthermore, the findings
correlate the findings published by other authors in this research area. Therefore, the findings
obtained from this study can be used by future investigators investigating in this area and
study of EGFP with different cell types might be useful for better understanding of the
properties of EGFP.

17LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
References:
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W., & Prasher, D. C. (1994). Green
fluorescent protein as a marker for gene expression. Science, 263(5148), 802-805.
Cody, C. W., Prasher, D. C., Westler, W. M., Prendergast, F. G., & Ward, W. W. (1993).
Chemical structure of the hexapeptide chromophore of the Aequorea green-
fluorescent protein. Biochemistry, 32(5), 1212-1218.
Cormack, B.P., Valdivia, R.H. and Falkow, S. (1996) ‘FACS-optimized mutants of the green
fluorescent protein (GFP)’, Gene, 173(1), pp. 33–38.
Lang, T., Goyard, S., Lebastard, M., & Milon, G. (2005). Bioluminescent Leishmania
expressing luciferase for rapid and high throughput screening of drugs acting on
amastigote‐harbouring macrophages and for quantitative real‐time monitoring of
parasitism features in living mice. Cellular microbiology, 7(3), 383-392.
Patterson, G., Day, R. N., & Piston, D. (2001). Fluorescent protein spectra. Journal of cell
science, 114(5), 837-838.
Prasher, D. C., Eckenrode, V. K., Ward, W. W., Prendergast, F. G., & Cormier, M. J. (1992).
Primary structure of the Aequorea victoria green-fluorescent protein. Gene, 111(2),
229-233.
RCSB Bank. (2019). RCSB PDB - 1GFL: STRUCTURE OF GREEN FLUORESCENT
PROTEIN. Retrieved from https://www.rcsb.org/3d-view/1GFL/1
Thomas, C. L., & Maule, A. J. (2000). Limitations on the use of fused green fluorescent
protein to investigate structure–function relationships for the cauliflower mosaic virus
movement protein. Journal of General Virology, 81(7), 1851-1855.
References:
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W., & Prasher, D. C. (1994). Green
fluorescent protein as a marker for gene expression. Science, 263(5148), 802-805.
Cody, C. W., Prasher, D. C., Westler, W. M., Prendergast, F. G., & Ward, W. W. (1993).
Chemical structure of the hexapeptide chromophore of the Aequorea green-
fluorescent protein. Biochemistry, 32(5), 1212-1218.
Cormack, B.P., Valdivia, R.H. and Falkow, S. (1996) ‘FACS-optimized mutants of the green
fluorescent protein (GFP)’, Gene, 173(1), pp. 33–38.
Lang, T., Goyard, S., Lebastard, M., & Milon, G. (2005). Bioluminescent Leishmania
expressing luciferase for rapid and high throughput screening of drugs acting on
amastigote‐harbouring macrophages and for quantitative real‐time monitoring of
parasitism features in living mice. Cellular microbiology, 7(3), 383-392.
Patterson, G., Day, R. N., & Piston, D. (2001). Fluorescent protein spectra. Journal of cell
science, 114(5), 837-838.
Prasher, D. C., Eckenrode, V. K., Ward, W. W., Prendergast, F. G., & Cormier, M. J. (1992).
Primary structure of the Aequorea victoria green-fluorescent protein. Gene, 111(2),
229-233.
RCSB Bank. (2019). RCSB PDB - 1GFL: STRUCTURE OF GREEN FLUORESCENT
PROTEIN. Retrieved from https://www.rcsb.org/3d-view/1GFL/1
Thomas, C. L., & Maule, A. J. (2000). Limitations on the use of fused green fluorescent
protein to investigate structure–function relationships for the cauliflower mosaic virus
movement protein. Journal of General Virology, 81(7), 1851-1855.

18LAB REPORT ON GREEN FLUORESCENT PROTEIN (GFP)
Tsien, R.Y. 1998. The green fluorescent protein. Annual review of biochemistry. 67(1),
pp.509544.
Zimmer, M. (2002). Green fluorescent protein (GFP): applications, structure, and related
photophysical behavior. Chemical reviews, 102(3), 759-782.
Tsien, R.Y. 1998. The green fluorescent protein. Annual review of biochemistry. 67(1),
pp.509544.
Zimmer, M. (2002). Green fluorescent protein (GFP): applications, structure, and related
photophysical behavior. Chemical reviews, 102(3), 759-782.
1 out of 19
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
© 2024 | Zucol Services PVT LTD | All rights reserved.