Laboratory Techniques for Gene Identification in Genetic Diseases
VerifiedAdded on 2022/09/21
|12
|2027
|40
Report
AI Summary
This report explores various laboratory techniques used for gene identification in the context of several diseases. It begins by detailing Fluorescence in situ hybridization (FISH) for identifying genes causing Chronic Myeloid Leukemia (CML) and Flow Cytometry for Acute Myeloid Leukemia (AML). The report then examines Restriction Fragment Length Polymorphism (RFLP) for sickle cell anemia, Allele-Specific PCR (ARMS) for B-Thalassemia, and Gap-PCR for alpha-Thalassemia, explaining the principles and applications of each technique. The report includes visual representations of positive and negative results for each method, aiding in understanding the diagnostic processes. Finally, it provides a comprehensive overview of how these techniques are used in molecular diagnostics to identify genetic mutations and diagnose hematological diseases.

Running head: LABORATORY TECHNIQUES
LABORATORY TECHNIQUES
Name of the Student
Name of the University
Author note
LABORATORY TECHNIQUES
Name of the Student
Name of the University
Author note
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

LABORATORY TECHNIQUES 1
Answer1
Genes causing Chronic myeloid leukaemia (CML) can be identified through FISH
(Fluorescence in situ hybridization). It is another way to look at the chromosome. Fluorescent
dye is used, which binds specifically to the genes or particular areas in the chromosome. In the
case of CML, FISH techniques is used to identify the specific part of BCR-ABL gene on
chromosome (5). The dye tagging makes the defective gene easily visible and hence diagnosis
can be done. It is mainly used in the bone marrow or blood samples without culturing in the cells
first.
In case of Acute Myeloid leukaemia (AML), genes can be identified with various
techniques/ The commonly used technique is flow cytometry. The progenitors are having a
common percentage of CD45-positive and non-erythroid events. Various denominator
determines the morphologic enumeration (8). Morphologic as well as differentiation dependant
arrangements of leukaemia are restricted in their prognostic importance; cytogenetics in addition
to molecular genetics look to be significant for classifying entities having distinct forecasts and
clinical behaviour. Hence, the genes can easily be classified.
RFLP techniques is used in identifying the genes for sickle cell anaemia. Person suffering
from the sickle cell mutation do not have restriction site for normal β-globin genes. The absence
of this gene causes sickle cell anemia. RLFP helps in identifying the missing genes. In RFLP
technique, uses radioactive labelled β-globin probe for analysing sickle cell anaemia. The
mutation that reasons this disease corresponds with a Cvn I site (6). Chromosomes which is
lacking or having sickle mutation do not have the site where the chromosomes normal β-globin
genes. The process of digestion with is over with Cvn I in addition to hybridization with a β-
globin probe, DNA from the patient with sickle-cell anaemia produces fragment of size 1.3 kb,
Answer1
Genes causing Chronic myeloid leukaemia (CML) can be identified through FISH
(Fluorescence in situ hybridization). It is another way to look at the chromosome. Fluorescent
dye is used, which binds specifically to the genes or particular areas in the chromosome. In the
case of CML, FISH techniques is used to identify the specific part of BCR-ABL gene on
chromosome (5). The dye tagging makes the defective gene easily visible and hence diagnosis
can be done. It is mainly used in the bone marrow or blood samples without culturing in the cells
first.
In case of Acute Myeloid leukaemia (AML), genes can be identified with various
techniques/ The commonly used technique is flow cytometry. The progenitors are having a
common percentage of CD45-positive and non-erythroid events. Various denominator
determines the morphologic enumeration (8). Morphologic as well as differentiation dependant
arrangements of leukaemia are restricted in their prognostic importance; cytogenetics in addition
to molecular genetics look to be significant for classifying entities having distinct forecasts and
clinical behaviour. Hence, the genes can easily be classified.
RFLP techniques is used in identifying the genes for sickle cell anaemia. Person suffering
from the sickle cell mutation do not have restriction site for normal β-globin genes. The absence
of this gene causes sickle cell anemia. RLFP helps in identifying the missing genes. In RFLP
technique, uses radioactive labelled β-globin probe for analysing sickle cell anaemia. The
mutation that reasons this disease corresponds with a Cvn I site (6). Chromosomes which is
lacking or having sickle mutation do not have the site where the chromosomes normal β-globin
genes. The process of digestion with is over with Cvn I in addition to hybridization with a β-
globin probe, DNA from the patient with sickle-cell anaemia produces fragment of size 1.3 kb,

LABORATORY TECHNIQUES 2
DNA received from individuals with two normal β-globin genes produces 1.1 kb fragment, in
addition to transporter having 1.1- and 1.3-kb fragments.
ARMS technique is used in gene identification causing B-Thalassemia. The process
depends on the theory of allele-specific priming of the PCR (polymerase chain reaction) method,
which is a detailed primer that allows amplification at 3’ terminal nucleotide matches with its
target sequence. β-thalassaemia occurring can be identified my mutation in gene IVSI-5 (G→C),
here the 3’ nucleotide of the ARMS primer is G to base pair through the substituted C in mutant
DNA. The primer lead to form G-G mismatch in the normal DNA however it is the discrepancy
occurring for a short time and hence would not cause any extension of the primer by itself (2).
This mismatching mutation would help in the identification of β-thalassaemia. The strong
mismatches such as C-C, G-A and A-A were observed for reducing the efficiency to zero or less
than 5% (8). Hence, it prevents further amplification, mismatch with target sequence that needs
to be added in the second, third and fourth nucleotide from 3’ end of the primer. Therefore, as
there is no extension in the gene amplification easy identification can be done.
Gene causing α-Thalassemia can be identified by gap- PCR. Most of the α-thalassemia’s
occurs due to obliteration of one or more HBA genes, besides these deletions are also common in
the population affected with the disease. The molecular diagnostic techniques mainly focused on
explaining and identifying the cause of the common deletion process (1). Gap-PCR are the
chosen technique in this discussion. PCR primers which can flank the mutual breakpoints have
the ability to demonstrate the deletion process by generation of the PCR products of particular
size. The certain primers are used in laboratories depending on the common deletion of the
population that are being served. Gap-PCR easily then detects deletion of the HBA gene which is
missed during DNA sequencing. It has also been noticed that the Gap-PCR that helps in the
DNA received from individuals with two normal β-globin genes produces 1.1 kb fragment, in
addition to transporter having 1.1- and 1.3-kb fragments.
ARMS technique is used in gene identification causing B-Thalassemia. The process
depends on the theory of allele-specific priming of the PCR (polymerase chain reaction) method,
which is a detailed primer that allows amplification at 3’ terminal nucleotide matches with its
target sequence. β-thalassaemia occurring can be identified my mutation in gene IVSI-5 (G→C),
here the 3’ nucleotide of the ARMS primer is G to base pair through the substituted C in mutant
DNA. The primer lead to form G-G mismatch in the normal DNA however it is the discrepancy
occurring for a short time and hence would not cause any extension of the primer by itself (2).
This mismatching mutation would help in the identification of β-thalassaemia. The strong
mismatches such as C-C, G-A and A-A were observed for reducing the efficiency to zero or less
than 5% (8). Hence, it prevents further amplification, mismatch with target sequence that needs
to be added in the second, third and fourth nucleotide from 3’ end of the primer. Therefore, as
there is no extension in the gene amplification easy identification can be done.
Gene causing α-Thalassemia can be identified by gap- PCR. Most of the α-thalassemia’s
occurs due to obliteration of one or more HBA genes, besides these deletions are also common in
the population affected with the disease. The molecular diagnostic techniques mainly focused on
explaining and identifying the cause of the common deletion process (1). Gap-PCR are the
chosen technique in this discussion. PCR primers which can flank the mutual breakpoints have
the ability to demonstrate the deletion process by generation of the PCR products of particular
size. The certain primers are used in laboratories depending on the common deletion of the
population that are being served. Gap-PCR easily then detects deletion of the HBA gene which is
missed during DNA sequencing. It has also been noticed that the Gap-PCR that helps in the
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
LABORATORY TECHNIQUES 3
diagnosis of α-thalassaemia are associated with the rare kind of mutations. Hence, different other
technique is being employed such as next generation sequencing (NGS) as well as Multiple
ligation-dependent probe amplification (MLPA) technologies that are used in order to
characterise the mutation that is occurring in an individual at the molecular level and causing the
disease. Research study have shown that the basic principal of this gap-pcr depends on the
inability of the primers for generating PCR product until and unless the deletions gets back to the
flanking sequence together.
Answer 2
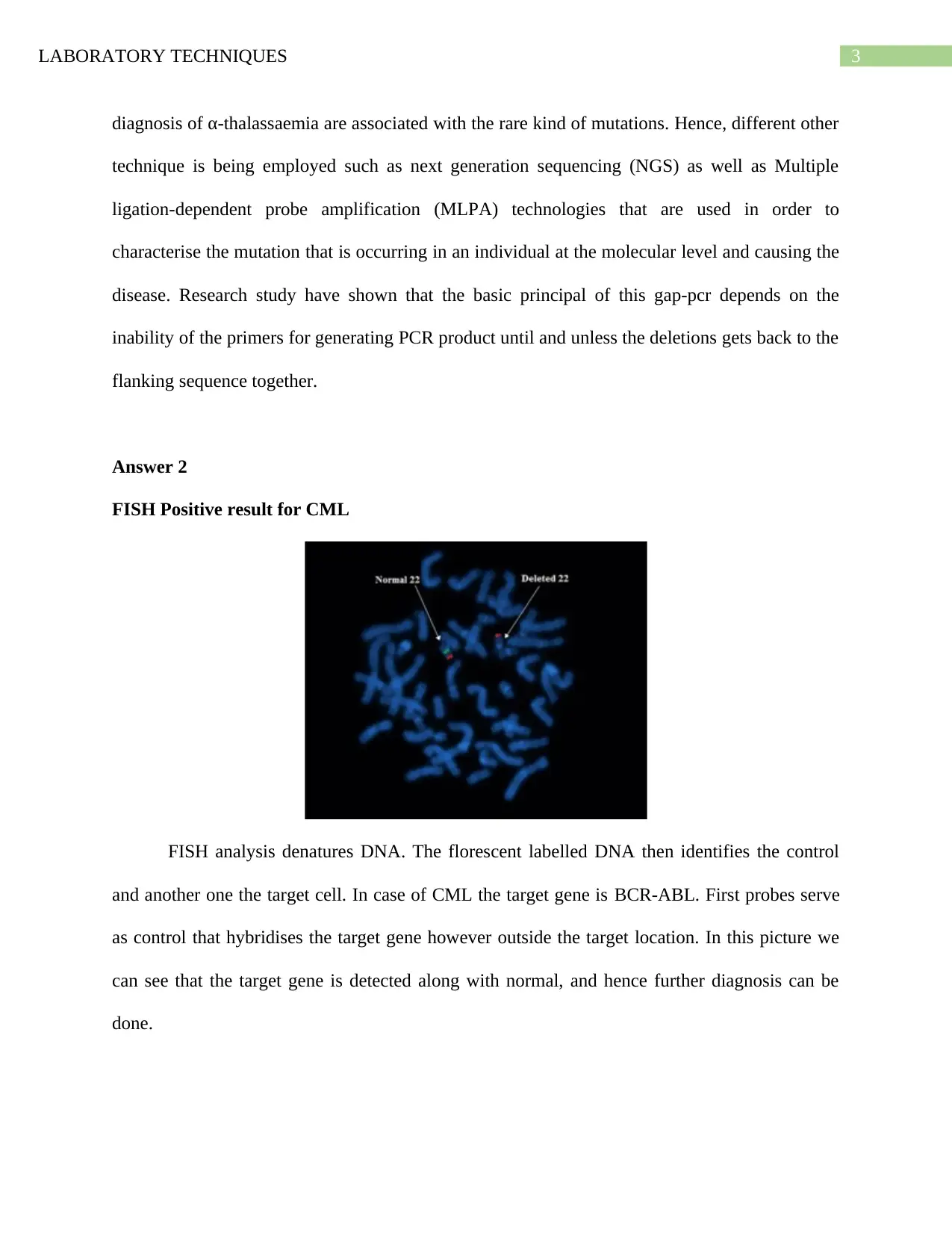
FISH Positive result for CML
FISH analysis denatures DNA. The florescent labelled DNA then identifies the control
and another one the target cell. In case of CML the target gene is BCR-ABL. First probes serve
as control that hybridises the target gene however outside the target location. In this picture we
can see that the target gene is detected along with normal, and hence further diagnosis can be
done.
diagnosis of α-thalassaemia are associated with the rare kind of mutations. Hence, different other
technique is being employed such as next generation sequencing (NGS) as well as Multiple
ligation-dependent probe amplification (MLPA) technologies that are used in order to
characterise the mutation that is occurring in an individual at the molecular level and causing the
disease. Research study have shown that the basic principal of this gap-pcr depends on the
inability of the primers for generating PCR product until and unless the deletions gets back to the
flanking sequence together.
Answer 2
FISH Positive result for CML
FISH analysis denatures DNA. The florescent labelled DNA then identifies the control
and another one the target cell. In case of CML the target gene is BCR-ABL. First probes serve
as control that hybridises the target gene however outside the target location. In this picture we
can see that the target gene is detected along with normal, and hence further diagnosis can be
done.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
LABORATORY TECHNIQUES 4
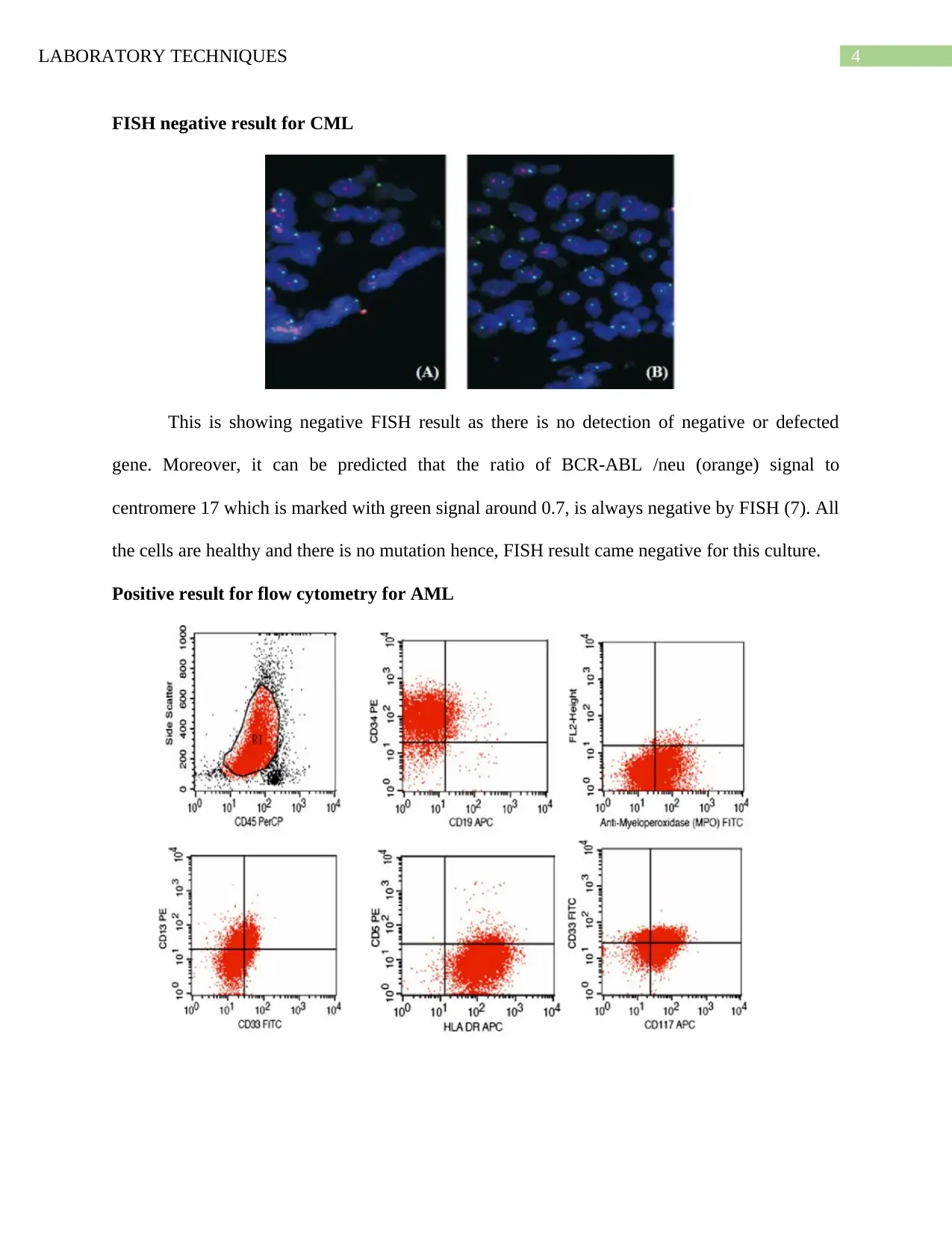
FISH negative result for CML
This is showing negative FISH result as there is no detection of negative or defected
gene. Moreover, it can be predicted that the ratio of BCR-ABL /neu (orange) signal to
centromere 17 which is marked with green signal around 0.7, is always negative by FISH (7). All
the cells are healthy and there is no mutation hence, FISH result came negative for this culture.
Positive result for flow cytometry for AML
FISH negative result for CML
This is showing negative FISH result as there is no detection of negative or defected
gene. Moreover, it can be predicted that the ratio of BCR-ABL /neu (orange) signal to
centromere 17 which is marked with green signal around 0.7, is always negative by FISH (7). All
the cells are healthy and there is no mutation hence, FISH result came negative for this culture.
Positive result for flow cytometry for AML
LABORATORY TECHNIQUES 5
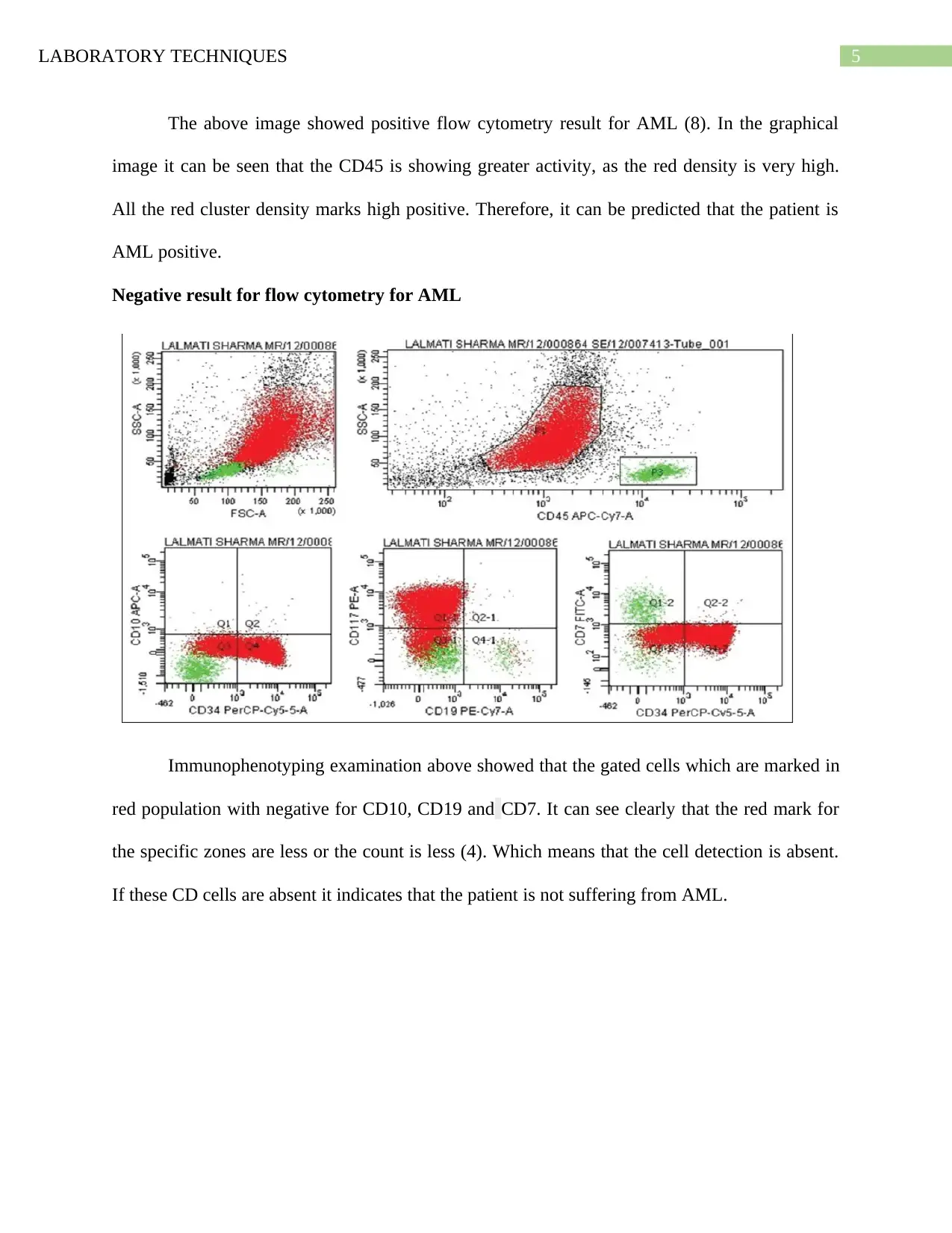
The above image showed positive flow cytometry result for AML (8). In the graphical
image it can be seen that the CD45 is showing greater activity, as the red density is very high.
All the red cluster density marks high positive. Therefore, it can be predicted that the patient is
AML positive.
Negative result for flow cytometry for AML
Immunophenotyping examination above showed that the gated cells which are marked in
red population with negative for CD10, CD19 and CD7. It can see clearly that the red mark for
the specific zones are less or the count is less (4). Which means that the cell detection is absent.
If these CD cells are absent it indicates that the patient is not suffering from AML.
The above image showed positive flow cytometry result for AML (8). In the graphical
image it can be seen that the CD45 is showing greater activity, as the red density is very high.
All the red cluster density marks high positive. Therefore, it can be predicted that the patient is
AML positive.
Negative result for flow cytometry for AML
Immunophenotyping examination above showed that the gated cells which are marked in
red population with negative for CD10, CD19 and CD7. It can see clearly that the red mark for
the specific zones are less or the count is less (4). Which means that the cell detection is absent.
If these CD cells are absent it indicates that the patient is not suffering from AML.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

LABORATORY TECHNIQUES 6
Positive result for RFLP-sickle cell anaemia
The above two images show the positive result for sickle cell anaemia when the test was
conducted with RFLP. The images showed that the disease blood is having a distinct band, this
analysis drawn the conclusion that the patient is having sickle cell anaemia (9). The southern blot
helps in identifying unhealthy gene/ protein by which disease prediction can be done. The size of
gene/protein for sickle cell anaemia is 1.42 kbp.
Negative RFLP result for sickle cell anaemia
This case shows that the person is the carrier but not infected. Hence it can be considered
as negative result. The healthy individual has only one DNA but carrier have 2 DNA band. So it
can be considered that even the test is negative as the person is not affected with sickle cell
anaemia however the person is the carrier of the disease (4).
Positive result for RFLP-sickle cell anaemia
The above two images show the positive result for sickle cell anaemia when the test was
conducted with RFLP. The images showed that the disease blood is having a distinct band, this
analysis drawn the conclusion that the patient is having sickle cell anaemia (9). The southern blot
helps in identifying unhealthy gene/ protein by which disease prediction can be done. The size of
gene/protein for sickle cell anaemia is 1.42 kbp.
Negative RFLP result for sickle cell anaemia
This case shows that the person is the carrier but not infected. Hence it can be considered
as negative result. The healthy individual has only one DNA but carrier have 2 DNA band. So it
can be considered that even the test is negative as the person is not affected with sickle cell
anaemia however the person is the carrier of the disease (4).
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
LABORATORY TECHNIQUES 7
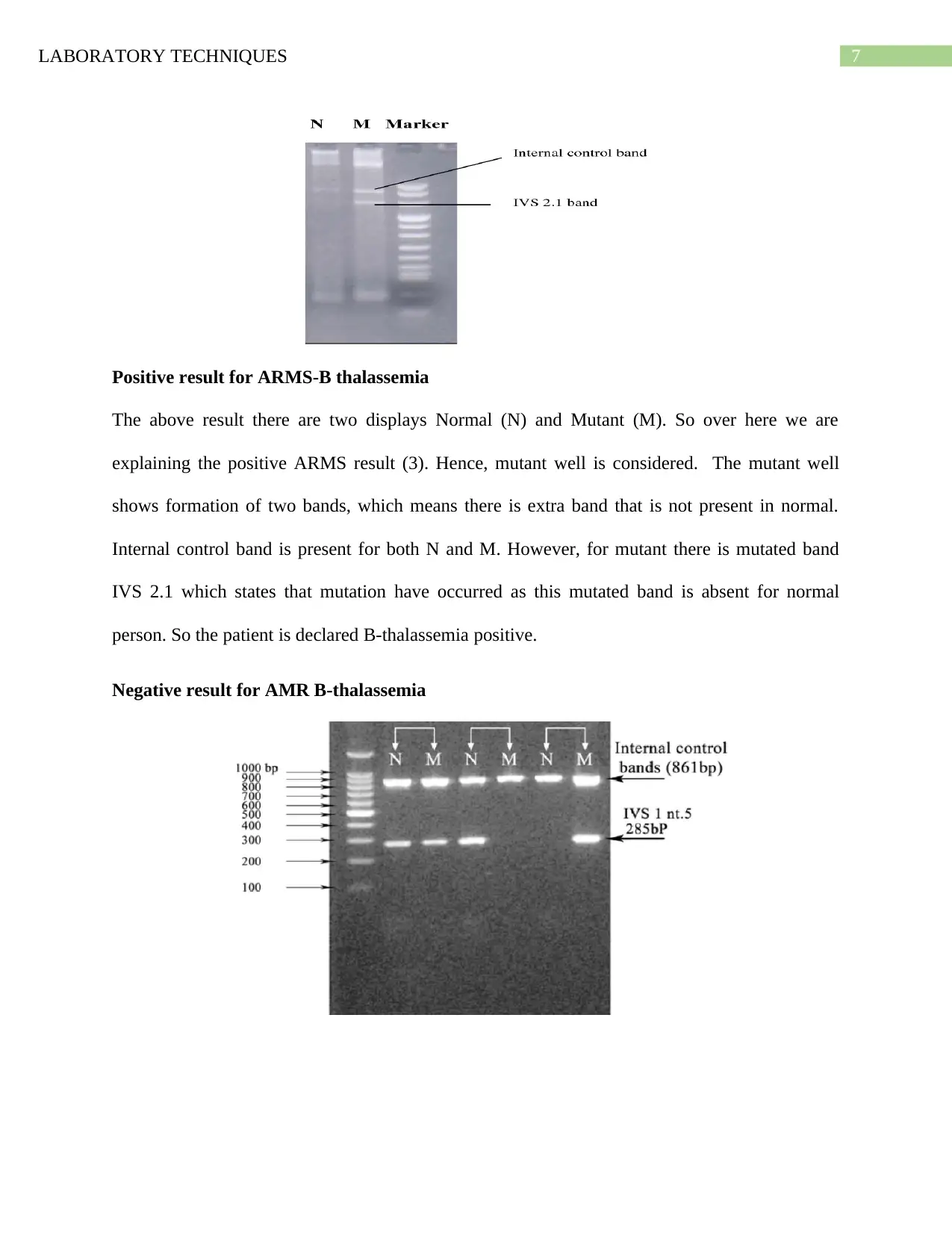
Positive result for ARMS-B thalassemia
The above result there are two displays Normal (N) and Mutant (M). So over here we are
explaining the positive ARMS result (3). Hence, mutant well is considered. The mutant well
shows formation of two bands, which means there is extra band that is not present in normal.
Internal control band is present for both N and M. However, for mutant there is mutated band
IVS 2.1 which states that mutation have occurred as this mutated band is absent for normal
person. So the patient is declared B-thalassemia positive.
Negative result for AMR B-thalassemia
Positive result for ARMS-B thalassemia
The above result there are two displays Normal (N) and Mutant (M). So over here we are
explaining the positive ARMS result (3). Hence, mutant well is considered. The mutant well
shows formation of two bands, which means there is extra band that is not present in normal.
Internal control band is present for both N and M. However, for mutant there is mutated band
IVS 2.1 which states that mutation have occurred as this mutated band is absent for normal
person. So the patient is declared B-thalassemia positive.
Negative result for AMR B-thalassemia
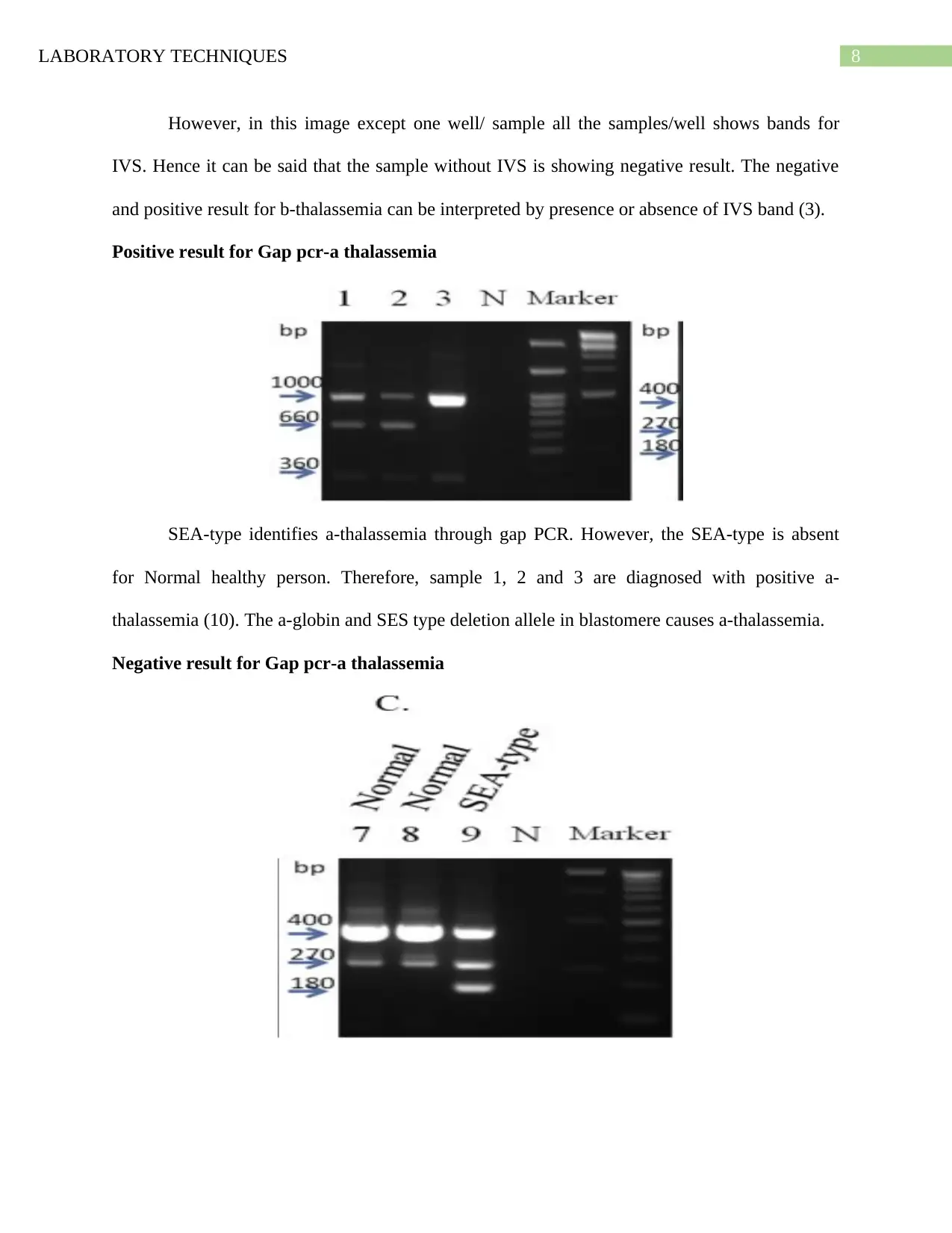
LABORATORY TECHNIQUES 8
However, in this image except one well/ sample all the samples/well shows bands for
IVS. Hence it can be said that the sample without IVS is showing negative result. The negative
and positive result for b-thalassemia can be interpreted by presence or absence of IVS band (3).
Positive result for Gap pcr-a thalassemia
SEA-type identifies a-thalassemia through gap PCR. However, the SEA-type is absent
for Normal healthy person. Therefore, sample 1, 2 and 3 are diagnosed with positive a-
thalassemia (10). The a-globin and SES type deletion allele in blastomere causes a-thalassemia.
Negative result for Gap pcr-a thalassemia
However, in this image except one well/ sample all the samples/well shows bands for
IVS. Hence it can be said that the sample without IVS is showing negative result. The negative
and positive result for b-thalassemia can be interpreted by presence or absence of IVS band (3).
Positive result for Gap pcr-a thalassemia
SEA-type identifies a-thalassemia through gap PCR. However, the SEA-type is absent
for Normal healthy person. Therefore, sample 1, 2 and 3 are diagnosed with positive a-
thalassemia (10). The a-globin and SES type deletion allele in blastomere causes a-thalassemia.
Negative result for Gap pcr-a thalassemia
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

LABORATORY TECHNIQUES 9
This picture demonstrates that 7 and 8 are normal they do not have SEA-type hence no
thalassemia. Hence, 7 and 8 are showing negative a-thalassemia result. There is no SEA band
other than 9. So 9 is positive and 7 and 8 are negative gap-pcr result (10).
This picture demonstrates that 7 and 8 are normal they do not have SEA-type hence no
thalassemia. Hence, 7 and 8 are showing negative a-thalassemia result. There is no SEA band
other than 9. So 9 is positive and 7 and 8 are negative gap-pcr result (10).
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

LABORATORY TECHNIQUES 10
References
1. Apsari NC. Pendampingan Bagi Anak Penyandang Thalasemia dan Keluarganya. Share:
Social Work Journal. 2016 Dec 24;6(2):154.
2. Fatmasyithah V, Rahayu MS. Gambaran Penderita Thalasemia Di Ruang Rawat Anak
Rumah Sakit Umum Cut Meutiaaceh Utara Tahun 2012. JESBIO: Jurnal Edukasi dan
Sains Biologi. 2016 Jan 13;3(2).
3. Furtado LF, Alves WP, Moreira TB, Junior LM, Miranda RR, Rabelo ÉM.
Standardization and application of the tetraprimer ARMS-PCR technique for screening of
the E198A SNP in the β-tubulin gene of hookworm populations in Brazil. Veterinary
parasitology. 2016 Jul 15;224:65-7.
4. Guida M, Cannavacciuolo PL, Cesarano M, Borra M, Biffali E, D’Alessandro R, De
Felice B. Microbial diversity of landslide soils assessed by RFLP and SSCP fingerprints.
Journal of applied genetics. 2014 Aug 1;55(3):403-15.
5. Höglund M, Sandin F, Simonsson B. Epidemiology of chronic myeloid leukaemia: an
update. Annals of hematology. 2015 Apr 1;94(2):241-7.
6. Kassim AA, Pruthi S, Day M, Rodeghier M, Gindville MC, Brodsky MA, DeBaun MR,
Jordan LC. Silent cerebral infarcts and cerebral aneurysms are prevalent in adults with
sickle cell anemia. Blood, The Journal of the American Society of Hematology. 2016 Apr
21;127(16):2038-40.
7. Liu G, Chen Z, Jiang X, Feng DQ, Zhao J, Fan D, Wang W. In-situ hydrothermal
synthesis of molecularly imprinted polymers coated carbon dots for fluorescent detection
of bisphenol A. Sensors and Actuators B: Chemical. 2016 Jun 2;228:302-7.
References
1. Apsari NC. Pendampingan Bagi Anak Penyandang Thalasemia dan Keluarganya. Share:
Social Work Journal. 2016 Dec 24;6(2):154.
2. Fatmasyithah V, Rahayu MS. Gambaran Penderita Thalasemia Di Ruang Rawat Anak
Rumah Sakit Umum Cut Meutiaaceh Utara Tahun 2012. JESBIO: Jurnal Edukasi dan
Sains Biologi. 2016 Jan 13;3(2).
3. Furtado LF, Alves WP, Moreira TB, Junior LM, Miranda RR, Rabelo ÉM.
Standardization and application of the tetraprimer ARMS-PCR technique for screening of
the E198A SNP in the β-tubulin gene of hookworm populations in Brazil. Veterinary
parasitology. 2016 Jul 15;224:65-7.
4. Guida M, Cannavacciuolo PL, Cesarano M, Borra M, Biffali E, D’Alessandro R, De
Felice B. Microbial diversity of landslide soils assessed by RFLP and SSCP fingerprints.
Journal of applied genetics. 2014 Aug 1;55(3):403-15.
5. Höglund M, Sandin F, Simonsson B. Epidemiology of chronic myeloid leukaemia: an
update. Annals of hematology. 2015 Apr 1;94(2):241-7.
6. Kassim AA, Pruthi S, Day M, Rodeghier M, Gindville MC, Brodsky MA, DeBaun MR,
Jordan LC. Silent cerebral infarcts and cerebral aneurysms are prevalent in adults with
sickle cell anemia. Blood, The Journal of the American Society of Hematology. 2016 Apr
21;127(16):2038-40.
7. Liu G, Chen Z, Jiang X, Feng DQ, Zhao J, Fan D, Wang W. In-situ hydrothermal
synthesis of molecularly imprinted polymers coated carbon dots for fluorescent detection
of bisphenol A. Sensors and Actuators B: Chemical. 2016 Jun 2;228:302-7.

LABORATORY TECHNIQUES 11
8. Podoltsev NA, Stahl M, Zeidan AM, Gore SD. Selecting initial treatment of acute
myeloid leukaemia in older adults. Blood reviews. 2017 Mar 1;31(2):43-62.
9. Vincent L, Vang D, Nguyen J, Benson B, Lei J, Gupta K. Cannabinoid receptor-specific
mechanisms to alleviate pain in sickle cell anemia via inhibition of mast cell activation
and neurogenic inflammation. Haematologica. 2016 May 1;101(5):566-77.
10. Yu LH, Liu D, Cai R, Shang X, Zhang XH, Ma XX, Yan SH, Fang P, Zheng CG, Wei
XF, Liu YH. Changes in hematological parameters in α‐thalassemia individuals co‐
inherited with erythroid Krüppel‐like factor mutations. Clinical genetics. 2015
Jul;88(1):56-61.
8. Podoltsev NA, Stahl M, Zeidan AM, Gore SD. Selecting initial treatment of acute
myeloid leukaemia in older adults. Blood reviews. 2017 Mar 1;31(2):43-62.
9. Vincent L, Vang D, Nguyen J, Benson B, Lei J, Gupta K. Cannabinoid receptor-specific
mechanisms to alleviate pain in sickle cell anemia via inhibition of mast cell activation
and neurogenic inflammation. Haematologica. 2016 May 1;101(5):566-77.
10. Yu LH, Liu D, Cai R, Shang X, Zhang XH, Ma XX, Yan SH, Fang P, Zheng CG, Wei
XF, Liu YH. Changes in hematological parameters in α‐thalassemia individuals co‐
inherited with erythroid Krüppel‐like factor mutations. Clinical genetics. 2015
Jul;88(1):56-61.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 12
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.