University Report on MALDI TOF MS: Background, Procedure, Results
VerifiedAdded on 2021/04/21
|15
|2809
|520
Report
AI Summary
This report provides a comprehensive overview of Matrix Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI TOF MS), a crucial technique in medical science. It begins with a historical background and explains the purpose and principles of MALDI TOF MS, emphasizing its role in identifying bacterial isolates through protein profiling. The report details the procedure, including sample preparation using Zip Tip, spotting techniques, and the use of the Vitek MS instrument. It also covers various applications such as bacteriology, virology, and mycology, along with a discussion of results, advantages (speed, accuracy, cost-effectiveness), and disadvantages (initial equipment cost, sensitivity limitations) of the technique. The report concludes with a list of references.

Running head: MALDI TOF MS
Medical Science
-MALDI TOF MS
Name of the Student
Name of the University
Author Note
Medical Science
-MALDI TOF MS
Name of the Student
Name of the University
Author Note
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

1MALDI TOF MS
Contents
MALDI TOF MS.......................................................................................................................2
Background and History:...........................................................................................................2
Purpose and Principles...............................................................................................................3
Procedure and Applications:......................................................................................................4
Name of the Instrument:.............................................................................................................4
How it works:.............................................................................................................................4
Reagents required:..................................................................................................................5
A. Processing Sample Using Zip Tip...............................................................................5
1. Equilibration............................................................................................................6
2. Loading....................................................................................................................6
3. Washing...................................................................................................................6
4. Elution......................................................................................................................6
B. Preparation of sample and spotting:............................................................................6
1. Preparation of matrix standard solution:..................................................................6
2. Standard spotting.....................................................................................................7
3. Sample spotting........................................................................................................7
Results:.......................................................................................................................................8
Advantages and Disadvantages:...............................................................................................10
Advantage:...........................................................................................................................10
Disadvantage:.......................................................................................................................10
References:...............................................................................................................................11
Contents
MALDI TOF MS.......................................................................................................................2
Background and History:...........................................................................................................2
Purpose and Principles...............................................................................................................3
Procedure and Applications:......................................................................................................4
Name of the Instrument:.............................................................................................................4
How it works:.............................................................................................................................4
Reagents required:..................................................................................................................5
A. Processing Sample Using Zip Tip...............................................................................5
1. Equilibration............................................................................................................6
2. Loading....................................................................................................................6
3. Washing...................................................................................................................6
4. Elution......................................................................................................................6
B. Preparation of sample and spotting:............................................................................6
1. Preparation of matrix standard solution:..................................................................6
2. Standard spotting.....................................................................................................7
3. Sample spotting........................................................................................................7
Results:.......................................................................................................................................8
Advantages and Disadvantages:...............................................................................................10
Advantage:...........................................................................................................................10
Disadvantage:.......................................................................................................................10
References:...............................................................................................................................11

2MALDI TOF MS
MALDI TOF MS
MALDI-TOF is an abbreviation for ‘Matrix Assisted Laser Desorption/Ionization
Time Of Flight Mass Spectrometer, which is a mass spectrometry technology that allows the
measurement of molecular mass of each atoms and compounds, converting them to charged
ions, and can be used to analyze biomolecules (Ru.ac.za, 2018). The technique involves an
ionization process called MALDI that utilizes a matrix that absorbs laser energy in a matrix
to create ions, without fragmentation of a large molecule and mass spectrophotometric
technique called TOF, which analyses the velocity of the created ions (recorded as the time of
flight) as per the mass to charge ratio of the ion to differentiate ions of different masses and
charges (Tuma, 2013).
Background and History:
The technique of matric assisted laser desoprption ionization (MALDI) was
developed by two Deutsche scientists: Michael Karas and Franz Hillencamp in 1985. They
found that alanine can be ionized more easily if mixed with tryptophan and then irradiated
with 266nm pulse. The tryptophan was able to absorb the energy from the radiation and
thereby ionize the non-absorbing alanine (Hillencamp & Karas, 2007). In 1987, Japanese
engineer, Koichi Tanaka showed that large proteins like carboxypeptidase-A can be ionized
by combining cobalt particles in glycerol and irradiating it with 337 nm nitrogen laser. This
proved that in the right setup, large protein molecules can be ionized easily (Sekiya et al.
2005). The time of flight Mass Spectrometer was first used by A.E. Cameron and D.F. Eggers
Jr. in 1948 (Katzenstein & Friedland, 1955; Mamyrin, 2001).
MALDI TOF MS
MALDI-TOF is an abbreviation for ‘Matrix Assisted Laser Desorption/Ionization
Time Of Flight Mass Spectrometer, which is a mass spectrometry technology that allows the
measurement of molecular mass of each atoms and compounds, converting them to charged
ions, and can be used to analyze biomolecules (Ru.ac.za, 2018). The technique involves an
ionization process called MALDI that utilizes a matrix that absorbs laser energy in a matrix
to create ions, without fragmentation of a large molecule and mass spectrophotometric
technique called TOF, which analyses the velocity of the created ions (recorded as the time of
flight) as per the mass to charge ratio of the ion to differentiate ions of different masses and
charges (Tuma, 2013).
Background and History:
The technique of matric assisted laser desoprption ionization (MALDI) was
developed by two Deutsche scientists: Michael Karas and Franz Hillencamp in 1985. They
found that alanine can be ionized more easily if mixed with tryptophan and then irradiated
with 266nm pulse. The tryptophan was able to absorb the energy from the radiation and
thereby ionize the non-absorbing alanine (Hillencamp & Karas, 2007). In 1987, Japanese
engineer, Koichi Tanaka showed that large proteins like carboxypeptidase-A can be ionized
by combining cobalt particles in glycerol and irradiating it with 337 nm nitrogen laser. This
proved that in the right setup, large protein molecules can be ionized easily (Sekiya et al.
2005). The time of flight Mass Spectrometer was first used by A.E. Cameron and D.F. Eggers
Jr. in 1948 (Katzenstein & Friedland, 1955; Mamyrin, 2001).
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

3MALDI TOF MS
The following section will discuss the purpose and principles of MALDI TOF,
followed by the procedure and application of the technique, discussion on typical resists
obtained from MALDI TOF analysis, and also a brief discussing on the advantages and
disadvantages of this technique.
Purpose and Principles
Propose of this technique is the identification of bacterial isolates.
MALDI TOF allows a fast identification of clinical bacterial isolates using
proteonomic based technique and protein profiling. This can be used as an alternative form of
other identification techniques to recognize microorganisms like gram positive bacteria,
Enterobacteriaceae, yeast, mold, non-fermenting bacteria and mycobacteria
(Schulthess et al., 2013; Conway et al., 2011; Blättel et al., 2013; Lau et al., 2012; Degand et
al., 2018; Panda et al., 2013).
The principle of this method is the identification of the plentiful proteins in the range
of 2 to 20kDa by analyzing their mass (m) to charge (z) ratio (m/z value). This helps in the
generation of a typical fingerprint for each type of microorganism, and can be used to
compare to a reference spectra to identify the sample. The principle is based on the
phenomenon of ionization of sample molecules when bombarded with laser.
This provides a straightforward, simple and quick technique for sample identification,
compared to genotypic and phenotypic processes (like immunological based techniques,
fluorescent in situ hybridization, Microarrays, DNA sequencing, Loop mediated isothermal
amplification, and metagenomic assay) (Panda et al., 2014). MALDI TOF allows
spectrometry of large biomolecules like proteins, and peptides are converted to ions by the
loss or addition of more than one proton. This is a ‘soft ionization’ process, which does not
The following section will discuss the purpose and principles of MALDI TOF,
followed by the procedure and application of the technique, discussion on typical resists
obtained from MALDI TOF analysis, and also a brief discussing on the advantages and
disadvantages of this technique.
Purpose and Principles
Propose of this technique is the identification of bacterial isolates.
MALDI TOF allows a fast identification of clinical bacterial isolates using
proteonomic based technique and protein profiling. This can be used as an alternative form of
other identification techniques to recognize microorganisms like gram positive bacteria,
Enterobacteriaceae, yeast, mold, non-fermenting bacteria and mycobacteria
(Schulthess et al., 2013; Conway et al., 2011; Blättel et al., 2013; Lau et al., 2012; Degand et
al., 2018; Panda et al., 2013).
The principle of this method is the identification of the plentiful proteins in the range
of 2 to 20kDa by analyzing their mass (m) to charge (z) ratio (m/z value). This helps in the
generation of a typical fingerprint for each type of microorganism, and can be used to
compare to a reference spectra to identify the sample. The principle is based on the
phenomenon of ionization of sample molecules when bombarded with laser.
This provides a straightforward, simple and quick technique for sample identification,
compared to genotypic and phenotypic processes (like immunological based techniques,
fluorescent in situ hybridization, Microarrays, DNA sequencing, Loop mediated isothermal
amplification, and metagenomic assay) (Panda et al., 2014). MALDI TOF allows
spectrometry of large biomolecules like proteins, and peptides are converted to ions by the
loss or addition of more than one proton. This is a ‘soft ionization’ process, which does not
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

4MALDI TOF MS
damage the structural integrity of the sample. The ions that are produced are then accelerated
in a fixed potential which differentiates them on the basis of their mass to charge ratios.
Various mass analyzers (like ion trap analyzer, quadrupole mass analyzer and time of flight
analyzer) to detect and measure these charged analytes. The determination of, the mass to
charge ratio is done by finding put the time for the charged ion to travel through the length of
the flight tube. The resultant information a peptide mass fingerprint (PMF) can be generated
for the analytes present in a sample (Singhal et al., 2015).
Procedure and Applications:
The MALDI TOF comprises of 3 parts: ion source, mass analyzer and a detector. The
MALDI (matrix) forms the source of the ion, while the flight tube and detector helps to detect
and analyze the ions (ru.ac.za, 2018).
Name of the Instrument:
Vitek MS- This is an automated identification system for microbes that uses mass
spectrometry technique and Matrix Assisted Desorption Ionization Time of Flight (MALDI-
TOF) technologies. The instrument contains a comprehensive CE marked as well as database
(for microbes) cleared by the FDA. The records include: accurate ID with Associated Spectra
Classifier, integrated ID/AST result and allows complete flexibility and traceability (VITEK®
MS, 2018).
How it works:
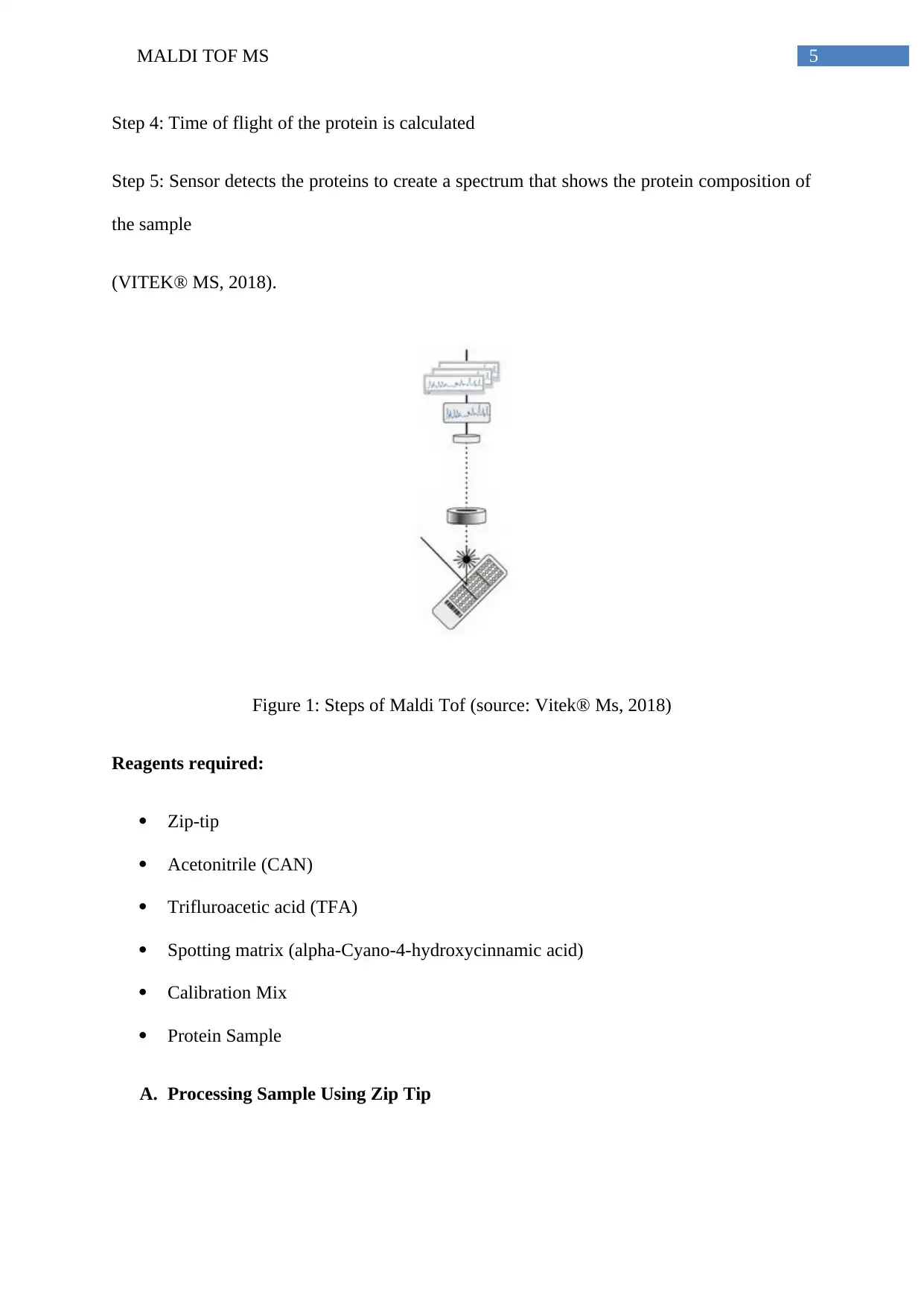
Step 1: preparation of target slide introduction into a high vacuum chamber
Step 2: Sample is ionized using laser
Step 3: Protein cloud released and accelerated due to the electric field
damage the structural integrity of the sample. The ions that are produced are then accelerated
in a fixed potential which differentiates them on the basis of their mass to charge ratios.
Various mass analyzers (like ion trap analyzer, quadrupole mass analyzer and time of flight
analyzer) to detect and measure these charged analytes. The determination of, the mass to
charge ratio is done by finding put the time for the charged ion to travel through the length of
the flight tube. The resultant information a peptide mass fingerprint (PMF) can be generated
for the analytes present in a sample (Singhal et al., 2015).
Procedure and Applications:
The MALDI TOF comprises of 3 parts: ion source, mass analyzer and a detector. The
MALDI (matrix) forms the source of the ion, while the flight tube and detector helps to detect
and analyze the ions (ru.ac.za, 2018).
Name of the Instrument:
Vitek MS- This is an automated identification system for microbes that uses mass
spectrometry technique and Matrix Assisted Desorption Ionization Time of Flight (MALDI-
TOF) technologies. The instrument contains a comprehensive CE marked as well as database
(for microbes) cleared by the FDA. The records include: accurate ID with Associated Spectra
Classifier, integrated ID/AST result and allows complete flexibility and traceability (VITEK®
MS, 2018).
How it works:
Step 1: preparation of target slide introduction into a high vacuum chamber
Step 2: Sample is ionized using laser
Step 3: Protein cloud released and accelerated due to the electric field
5MALDI TOF MS
Step 4: Time of flight of the protein is calculated
Step 5: Sensor detects the proteins to create a spectrum that shows the protein composition of
the sample
(VITEK® MS, 2018).
Figure 1: Steps of Maldi Tof (source: Vitek® Ms, 2018)
Reagents required:
Zip-tip
Acetonitrile (CAN)
Trifluroacetic acid (TFA)
Spotting matrix (alpha-Cyano-4-hydroxycinnamic acid)
Calibration Mix
Protein Sample
A. Processing Sample Using Zip Tip
Step 4: Time of flight of the protein is calculated
Step 5: Sensor detects the proteins to create a spectrum that shows the protein composition of
the sample
(VITEK® MS, 2018).
Figure 1: Steps of Maldi Tof (source: Vitek® Ms, 2018)
Reagents required:
Zip-tip
Acetonitrile (CAN)
Trifluroacetic acid (TFA)
Spotting matrix (alpha-Cyano-4-hydroxycinnamic acid)
Calibration Mix
Protein Sample
A. Processing Sample Using Zip Tip
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

6MALDI TOF MS
1. Equilibration: The Zip Tip is activated with 10 microlitre of acetonitrile (CAN)
thrice.
2. Loading: The sample is loaded to the Zip Tip by pipetting the sample (5 to 10
microlitre at a time) repeated 15 times, and then the rest of the liquid is discarded.
3. Washing: Salts are removed by washing with 3x10 microlitre of 10% TFA the
C18/C4 tip.
4. Elution: Sample is eluted from the Zip tip using 50% CAN in 0.1% TFA or directly
into the matrix (like CHCA in 70% ACN/0.1% TFA).
B. Preparation of sample and spotting:
1. Preparation of matrix standard solution:
The correct matrix is selected depending upon the molecular weight of the protein.
Matrix Selection:
Sample Matrix
Peptide less than 10
kDa
α-Cyano-4-hydroxycinnamic acid(CHCA)
Protein less than 10 kDa a. Sinapinic acid b. Super DHB
Polymer a. α-Cyano-4-hydroxycinnamic acid(CHCA)
b. 2,5 dihydroxybenzoic acid(DHB)
Glycosylated Protein Super DHB
The matrix solution is prepared in an a proper solvent [5 mg of alpha-cyano in
total of 0.5 mL solution containing 0.2 mL of 0.1% TFA and 0.3 mL of 100%
ACN]
1. Equilibration: The Zip Tip is activated with 10 microlitre of acetonitrile (CAN)
thrice.
2. Loading: The sample is loaded to the Zip Tip by pipetting the sample (5 to 10
microlitre at a time) repeated 15 times, and then the rest of the liquid is discarded.
3. Washing: Salts are removed by washing with 3x10 microlitre of 10% TFA the
C18/C4 tip.
4. Elution: Sample is eluted from the Zip tip using 50% CAN in 0.1% TFA or directly
into the matrix (like CHCA in 70% ACN/0.1% TFA).
B. Preparation of sample and spotting:
1. Preparation of matrix standard solution:
The correct matrix is selected depending upon the molecular weight of the protein.
Matrix Selection:
Sample Matrix
Peptide less than 10
kDa
α-Cyano-4-hydroxycinnamic acid(CHCA)
Protein less than 10 kDa a. Sinapinic acid b. Super DHB
Polymer a. α-Cyano-4-hydroxycinnamic acid(CHCA)
b. 2,5 dihydroxybenzoic acid(DHB)
Glycosylated Protein Super DHB
The matrix solution is prepared in an a proper solvent [5 mg of alpha-cyano in
total of 0.5 mL solution containing 0.2 mL of 0.1% TFA and 0.3 mL of 100%
ACN]
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

7MALDI TOF MS
The standard solution or premix is prepared by adding 10 microlitre of each
protein or peptide (10 picomoles per microlitre).
2. Standard spotting
0.5 microlitre of the matrix solution is deposited into the spot plate and left for
10s, and the excess amount is then removed
0.5 microliotre of pepmix is added to the matrix solution and 0.5 microlitre of
the matrix solution is then added to the sample (this is called the sandwich
method). The step is repeated for other spots, so that each standard spot is
surrounded with sample spots.
The dish is kept in the drier for 30mins, until the spots are dry and have a
uniform appearance (slightly yellow to off white color).
3. Sample spotting
0.5 microlitre of matrix solution is added to the spot and left for 10 sec and the
any remaining, additional solution is removed
0.5 microlitre of the sample is added to the matrix solution and then 0.5
microlitre of the matrix is added back to the sample. The step is repeated for
other samples, next to standard spot.
The plate is kept in the drier, and the dried matrix ought to have a uniform
look (slightly yellow to off white color).
For further tracking, the spot positions (for standard and samples) are
recorded.
The plate is then inserted in the MALDI equipment (Iitb.vlab.co.in, 2018).
Applications: Bacteriology (detection of food and water borne bacteria,
environmental bacteriology, detection and identification of bio-weapons, detecting
and identifying antibiotic resistance in bacteria, bacterial strain typing and taxonomy),
The standard solution or premix is prepared by adding 10 microlitre of each
protein or peptide (10 picomoles per microlitre).
2. Standard spotting
0.5 microlitre of the matrix solution is deposited into the spot plate and left for
10s, and the excess amount is then removed
0.5 microliotre of pepmix is added to the matrix solution and 0.5 microlitre of
the matrix solution is then added to the sample (this is called the sandwich
method). The step is repeated for other spots, so that each standard spot is
surrounded with sample spots.
The dish is kept in the drier for 30mins, until the spots are dry and have a
uniform appearance (slightly yellow to off white color).
3. Sample spotting
0.5 microlitre of matrix solution is added to the spot and left for 10 sec and the
any remaining, additional solution is removed
0.5 microlitre of the sample is added to the matrix solution and then 0.5
microlitre of the matrix is added back to the sample. The step is repeated for
other samples, next to standard spot.
The plate is kept in the drier, and the dried matrix ought to have a uniform
look (slightly yellow to off white color).
For further tracking, the spot positions (for standard and samples) are
recorded.
The plate is then inserted in the MALDI equipment (Iitb.vlab.co.in, 2018).
Applications: Bacteriology (detection of food and water borne bacteria,
environmental bacteriology, detection and identification of bio-weapons, detecting
and identifying antibiotic resistance in bacteria, bacterial strain typing and taxonomy),
8MALDI TOF MS
virology (clinical virology, viral genotyping and epidemiological studies), mycology
(clinical mycology, detecting antibiotic resistance in fungi, fungal strain typing)
(Singhal et al., 2015).
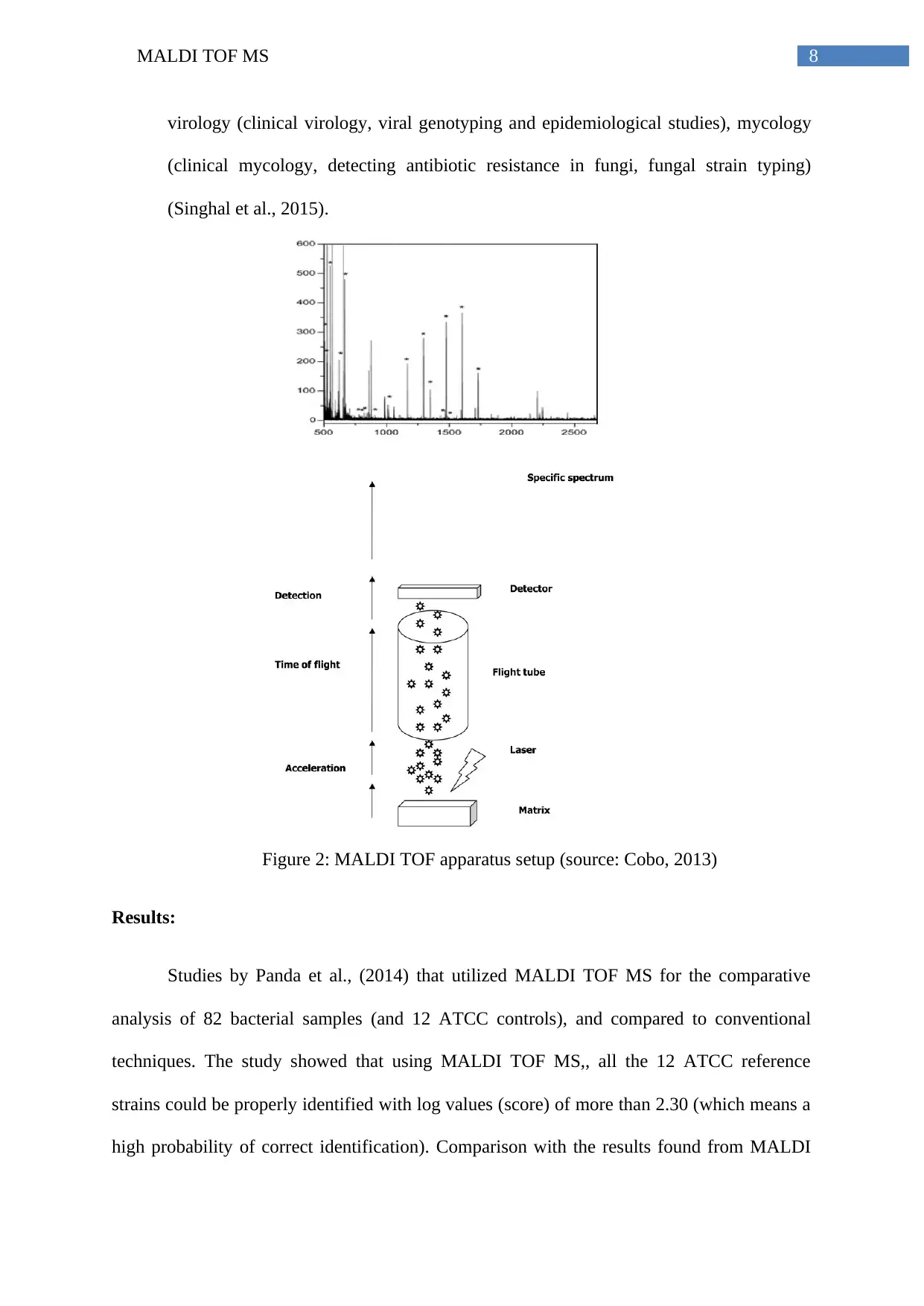
Figure 2: MALDI TOF apparatus setup (source: Cobo, 2013)
Results:
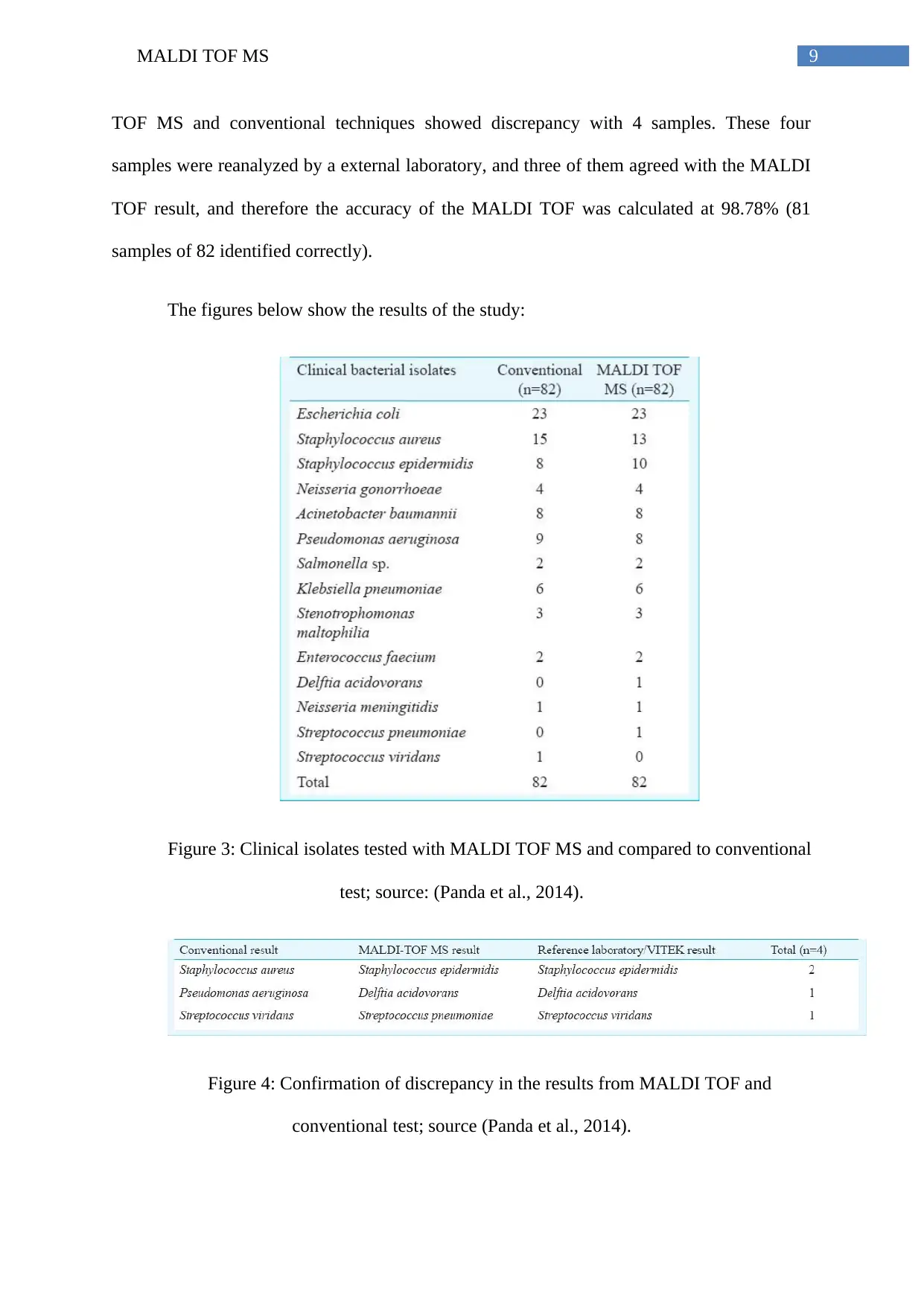
Studies by Panda et al., (2014) that utilized MALDI TOF MS for the comparative
analysis of 82 bacterial samples (and 12 ATCC controls), and compared to conventional
techniques. The study showed that using MALDI TOF MS,, all the 12 ATCC reference
strains could be properly identified with log values (score) of more than 2.30 (which means a
high probability of correct identification). Comparison with the results found from MALDI
virology (clinical virology, viral genotyping and epidemiological studies), mycology
(clinical mycology, detecting antibiotic resistance in fungi, fungal strain typing)
(Singhal et al., 2015).
Figure 2: MALDI TOF apparatus setup (source: Cobo, 2013)
Results:
Studies by Panda et al., (2014) that utilized MALDI TOF MS for the comparative
analysis of 82 bacterial samples (and 12 ATCC controls), and compared to conventional
techniques. The study showed that using MALDI TOF MS,, all the 12 ATCC reference
strains could be properly identified with log values (score) of more than 2.30 (which means a
high probability of correct identification). Comparison with the results found from MALDI
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
9MALDI TOF MS
TOF MS and conventional techniques showed discrepancy with 4 samples. These four
samples were reanalyzed by a external laboratory, and three of them agreed with the MALDI
TOF result, and therefore the accuracy of the MALDI TOF was calculated at 98.78% (81
samples of 82 identified correctly).
The figures below show the results of the study:
Figure 3: Clinical isolates tested with MALDI TOF MS and compared to conventional
test; source: (Panda et al., 2014).
Figure 4: Confirmation of discrepancy in the results from MALDI TOF and
conventional test; source (Panda et al., 2014).
TOF MS and conventional techniques showed discrepancy with 4 samples. These four
samples were reanalyzed by a external laboratory, and three of them agreed with the MALDI
TOF result, and therefore the accuracy of the MALDI TOF was calculated at 98.78% (81
samples of 82 identified correctly).
The figures below show the results of the study:
Figure 3: Clinical isolates tested with MALDI TOF MS and compared to conventional
test; source: (Panda et al., 2014).
Figure 4: Confirmation of discrepancy in the results from MALDI TOF and
conventional test; source (Panda et al., 2014).
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

10MALDI TOF MS
Advantages and Disadvantages:
Advantage:
It is fast, accurate, less expensive (than immunological based detection) and does not
require trained personnel (Singhal et al., 2015). Wide variety of specimen can be
characterized. The method is highly sensitive, providing high throughput and easy
preparation of sample. The process can also be automated and helps to improve patient
management (Cobo, 2013). It allows observation of ionized molecules without causing any
fragmentation since the ions have low internal energy. It can help in the identification of
microbes at the subspecies level (Augulis, 2018)
Disadvantage:
Initial cost of the equipment is high (Singhal et al. 2015). The process can also be
time consuming, and can involve different genetic markers (Cobo, 2013). The sensitivity is
low without a prior culture. The technique cannot detect a low amount of microbes in sterile
samples (Augulis, 2018).
Figure 5: Comparison of advantages and disadvantages of MALDI TOF (source:
Cobo, 2013).
Advantages and Disadvantages:
Advantage:
It is fast, accurate, less expensive (than immunological based detection) and does not
require trained personnel (Singhal et al., 2015). Wide variety of specimen can be
characterized. The method is highly sensitive, providing high throughput and easy
preparation of sample. The process can also be automated and helps to improve patient
management (Cobo, 2013). It allows observation of ionized molecules without causing any
fragmentation since the ions have low internal energy. It can help in the identification of
microbes at the subspecies level (Augulis, 2018)
Disadvantage:
Initial cost of the equipment is high (Singhal et al. 2015). The process can also be
time consuming, and can involve different genetic markers (Cobo, 2013). The sensitivity is
low without a prior culture. The technique cannot detect a low amount of microbes in sterile
samples (Augulis, 2018).
Figure 5: Comparison of advantages and disadvantages of MALDI TOF (source:
Cobo, 2013).

11MALDI TOF MS
References:
Augulis, R. (2018). What are advantages and disadvantages of MALDI Imaging mass
spectrometry vs next-generation sequencing in pathology research?. researchgate.net.
Retrieved 3 March 2018, from
https://www.researchgate.net/post/What_are_advantages_and_disadvantages_of_MA
LDI_Imaging_mass_spectrometry_vs_next-
generation_sequencing_in_pathology_research
Blättel, V., Petri, A., Rabenstein, A., Kuever, J., & König, H. (2013). Differentiation of
species of the genus Saccharomyces using biomolecular fingerprinting
methods. Applied microbiology and biotechnology, 97(10), 4597-4606.
Cobo, F. (2013). Application of MALDI-TOF Mass Spectrometry in Clinical Virology: A
Review. The Open Virology Journal, 7(1), 84-90.
http://dx.doi.org/10.2174/1874357920130927003
Conway, G. C., Smole, S. C., Sarracino, D. A., Arbeit, R. D., & Leopold, P. E. (2011).
Phyloproteomics: species identification of Enterobacteriaceae using matrix-assisted
laser desorption/ionization time-of-flight mass spectrometry. Journal of molecular
microbiology and biotechnology, 3(1), 103-112.
Degand, N., Carbonnelle, E., Dauphin, B., Beretti, J. L., Le Bourgeois, M., Sermet-Gaudelus,
I., ... & Ferroni, A. (2018). Matrix-assisted laser desorption ionization-time of flight
mass spectrometry for identification of nonfermenting gram-negative bacilli isolated
from cystic fibrosis patients. Journal of clinical microbiology, 46(10), 3361-3367.
Hillenkamp, F., & Karas, M. (2007). The MALDI process and method (pp. 1-28). Wiley‐
VCH Verlag GmbH & Co. KGaA.
References:
Augulis, R. (2018). What are advantages and disadvantages of MALDI Imaging mass
spectrometry vs next-generation sequencing in pathology research?. researchgate.net.
Retrieved 3 March 2018, from
https://www.researchgate.net/post/What_are_advantages_and_disadvantages_of_MA
LDI_Imaging_mass_spectrometry_vs_next-
generation_sequencing_in_pathology_research
Blättel, V., Petri, A., Rabenstein, A., Kuever, J., & König, H. (2013). Differentiation of
species of the genus Saccharomyces using biomolecular fingerprinting
methods. Applied microbiology and biotechnology, 97(10), 4597-4606.
Cobo, F. (2013). Application of MALDI-TOF Mass Spectrometry in Clinical Virology: A
Review. The Open Virology Journal, 7(1), 84-90.
http://dx.doi.org/10.2174/1874357920130927003
Conway, G. C., Smole, S. C., Sarracino, D. A., Arbeit, R. D., & Leopold, P. E. (2011).
Phyloproteomics: species identification of Enterobacteriaceae using matrix-assisted
laser desorption/ionization time-of-flight mass spectrometry. Journal of molecular
microbiology and biotechnology, 3(1), 103-112.
Degand, N., Carbonnelle, E., Dauphin, B., Beretti, J. L., Le Bourgeois, M., Sermet-Gaudelus,
I., ... & Ferroni, A. (2018). Matrix-assisted laser desorption ionization-time of flight
mass spectrometry for identification of nonfermenting gram-negative bacilli isolated
from cystic fibrosis patients. Journal of clinical microbiology, 46(10), 3361-3367.
Hillenkamp, F., & Karas, M. (2007). The MALDI process and method (pp. 1-28). Wiley‐
VCH Verlag GmbH & Co. KGaA.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 15
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.




