Microbiology and Biochemistry Report - Laboratory Analysis
VerifiedAdded on 2020/05/04
|18
|3640
|253
Report
AI Summary
This microbiology and biochemistry report covers several key aspects of laboratory techniques and bacterial analysis. It begins with aseptic techniques, detailing precautions to prevent contamination. The report then explores the streak plate method, explaining its purpose and procedure. It also examines Escherichia coli, including its habitat, infections caused, and the significance of its strains. The report further discusses selective and differential growth media, with a focus on MacConkey agar and the properties of E. coli colonies. Contamination sources and prevention methods are addressed, followed by a comparison of Gram-positive and Gram-negative stains and the expected appearance of E. coli and Bacillus subtilis under a microscope. The report contrasts streak and spread plate techniques and discusses viable plate counts. Additionally, it delves into Yakult, explaining its probiotic nature and the species it contains. The report also analyzes colony forming units (CFU) and relates it to the advertised quantity of bacteria in Yakult. Finally, the report covers bacterial identification using API 20E test strips and 16S rRNA gene sequencing, discussing the significance of identified species and advantages of the sequencing method.

Microbiology and biochemistry
Name
Name of the class
Instructor
Institution
Date
No. Of words: 3026
Name
Name of the class
Instructor
Institution
Date
No. Of words: 3026
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

1. Aseptic technique
Aseptic technique is defined as a laboratory skill used by microbiologists for various procedures
including isolation of pure cultures, executing microbiological tests and transferring cultures
among other functions. The technique prevents contamination of cultures from bacteria found in
the environment like the microbes found on the researchers body that might contaminate
cultures, hence interfering with the actual laboratory results. Besides, aseptic technique can also
be used to isolate individual species of microorganisms from a mixed culture so as to get a new
culture.
2. Necessary precautions that ensure aseptic technique especially when manipulating specimens
in a laboratory.
There are a myriad of factors that should be taken for proper aseptic method especially when
manipulation samples in the lab and they include; The first precaution is to ensure that you avoid
cross contamination by working with one microorganism at a time. Switching of cell lines or
microorganism either intentionally or accidentally do occur most of the times which can lead to
misleading or erroneous information
Second, work areas and biological safety cabinet’s needs to be thoroughly cleaned as well as
disinfected between vulture sessions. It is also recommended to avoid routine antibiotics
especially during cell culture media, because antibiotics can cover the underlying contamination
that is not of target by antibiotics.
Above all, the most basic procedures include using proper antiseptic procedure, wearing clean
dust coats and thoroughly washing of hands to ensure microorganisms are introduced into the
mammalian cell cultures
Streak plate method
1. The need for streaking bacterium on growth media with a loop
The aim of streaking bacterium is to isolate or define a given colony from mixed population
although streaking is ideal for separating, it does not work with larger volumes neither does it
give the concentration count.
Aseptic technique is defined as a laboratory skill used by microbiologists for various procedures
including isolation of pure cultures, executing microbiological tests and transferring cultures
among other functions. The technique prevents contamination of cultures from bacteria found in
the environment like the microbes found on the researchers body that might contaminate
cultures, hence interfering with the actual laboratory results. Besides, aseptic technique can also
be used to isolate individual species of microorganisms from a mixed culture so as to get a new
culture.
2. Necessary precautions that ensure aseptic technique especially when manipulating specimens
in a laboratory.
There are a myriad of factors that should be taken for proper aseptic method especially when
manipulation samples in the lab and they include; The first precaution is to ensure that you avoid
cross contamination by working with one microorganism at a time. Switching of cell lines or
microorganism either intentionally or accidentally do occur most of the times which can lead to
misleading or erroneous information
Second, work areas and biological safety cabinet’s needs to be thoroughly cleaned as well as
disinfected between vulture sessions. It is also recommended to avoid routine antibiotics
especially during cell culture media, because antibiotics can cover the underlying contamination
that is not of target by antibiotics.
Above all, the most basic procedures include using proper antiseptic procedure, wearing clean
dust coats and thoroughly washing of hands to ensure microorganisms are introduced into the
mammalian cell cultures
Streak plate method
1. The need for streaking bacterium on growth media with a loop
The aim of streaking bacterium is to isolate or define a given colony from mixed population
although streaking is ideal for separating, it does not work with larger volumes neither does it
give the concentration count.

2. How streak plate technique is done as well as when the loop is sterilized or when to use a fresh
sterile
Start by cooling your loop by touching it on the sides of the agar plate. Next, put the nichrome
wire into a culture of the bacteria mixture. Pull the lid off your plate, insert the nichrome wire
and then drag it over its top surface. This should be done in a back and forth zigzag motion as
shown in the diagram below.
Escherichia coli
1. What’s the natural habitat for Escherichia coli?
The main habitat of this bacterium is in the gut of organisms (both animals and people)
2. The range of infections that are caused by strains of Escherichia coli?
The main infections are gastroenteritis, urinary tract infections, gut infections with severe bloody
diarrhea, intra abdominal infections. Other infections are meningitis, pneumonia, infected
bones/joints, and infections of the skin or tissues.
3. All strains of Escherichia coli are not equally able to cause infections or diseases because they
are harmless strains that live in the human colon and they are of benefit since they produce
vitamin K which helps in synthesis of proteins in the body.
sterile
Start by cooling your loop by touching it on the sides of the agar plate. Next, put the nichrome
wire into a culture of the bacteria mixture. Pull the lid off your plate, insert the nichrome wire
and then drag it over its top surface. This should be done in a back and forth zigzag motion as
shown in the diagram below.
Escherichia coli
1. What’s the natural habitat for Escherichia coli?
The main habitat of this bacterium is in the gut of organisms (both animals and people)
2. The range of infections that are caused by strains of Escherichia coli?
The main infections are gastroenteritis, urinary tract infections, gut infections with severe bloody
diarrhea, intra abdominal infections. Other infections are meningitis, pneumonia, infected
bones/joints, and infections of the skin or tissues.
3. All strains of Escherichia coli are not equally able to cause infections or diseases because they
are harmless strains that live in the human colon and they are of benefit since they produce
vitamin K which helps in synthesis of proteins in the body.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

4. The property of Escherichia coli strain used for the untrained microbiologists especially in the
first year practical is virulence properties
5. The reason to why plates were inoculated with presumed Escherichia coli incubated at 37
degrees Celsius was to promote the growth of target group of E coli
Selective & differential growth media
1. Define the terms differential and selective growth media
For the selective growth media, it allows specific type of species to thrive as well as hinders the
growth of other species while the differential growth media applies when distinguishing closely
related animals.
2. The likelihood appearance of Escherichia coli growing on MacConkey agar is the colonies
that are pinkish to red in color and a bile salt precipitate surrounding the colonies.
3. What property does the colour of E. coli growing on MacConkey agar indicate?
The property is virulence.
Contamination
1. The reason to why you expect multiple contaminants to thrive on nutrient agar plates other
than MacConkey agar is due to the subtype of nutrient agar which is the main medium for
microbial studies. Besides, it can also be used during routine cultivation of species. Also, the
nutrient agar plate does not grow one type of bacteria over the other. On the other hand,
MacConkey is an agar where the Gram negative grows. What is more is that Escherichia coli
grow in red colonies because of the PH indicator. Also, it is important to mention that
MacConkey agar powder is of two forms; one with carbohydrate lactose in it plus the other with
no added sugar and because Escherichia coli ferment carbohydrates to acids, you can put
different forms of sugar to the sugar free MacConkey agar to check whether red colonies will
develop. In case they develop, it means that the Escherichia coli strain being used can use such
sugar
first year practical is virulence properties
5. The reason to why plates were inoculated with presumed Escherichia coli incubated at 37
degrees Celsius was to promote the growth of target group of E coli
Selective & differential growth media
1. Define the terms differential and selective growth media
For the selective growth media, it allows specific type of species to thrive as well as hinders the
growth of other species while the differential growth media applies when distinguishing closely
related animals.
2. The likelihood appearance of Escherichia coli growing on MacConkey agar is the colonies
that are pinkish to red in color and a bile salt precipitate surrounding the colonies.
3. What property does the colour of E. coli growing on MacConkey agar indicate?
The property is virulence.
Contamination
1. The reason to why you expect multiple contaminants to thrive on nutrient agar plates other
than MacConkey agar is due to the subtype of nutrient agar which is the main medium for
microbial studies. Besides, it can also be used during routine cultivation of species. Also, the
nutrient agar plate does not grow one type of bacteria over the other. On the other hand,
MacConkey is an agar where the Gram negative grows. What is more is that Escherichia coli
grow in red colonies because of the PH indicator. Also, it is important to mention that
MacConkey agar powder is of two forms; one with carbohydrate lactose in it plus the other with
no added sugar and because Escherichia coli ferment carbohydrates to acids, you can put
different forms of sugar to the sugar free MacConkey agar to check whether red colonies will
develop. In case they develop, it means that the Escherichia coli strain being used can use such
sugar
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

2. The possible sources or contamination and how they can be avoided in further laboratory
experiments
The common sources of contamination include contamination from analysts, low quality
reagents, analysts, visitor’s entry, sampling tools, sample environment and container. To prevent
such errors, it would be ideal reagents plus quality of recommended grades from reputable
dealers. One should also maintain the recommended condition between analysis and the sample
collection. It is also important to use clean glassware during analysis and ensure care as per the
recommendations in the microbial testing labs.
Gram stain
1. The main difference between gram positive and gram negative stain is that in gram positive
cell walls have a thick layer of techoic acids referred to as peptidoglycan, while in the gram
negative bacteria, the cell walls have a thin layer without techoic acids. Besides, gram positive
bacteria do stain purple due to the thick layer mentioned above while gram negative bacteria do
stain red due to the thin peptidoglycan layer that is surrounded by the plasma membrane and
therefore will not stain with crystal violet
2. The expected appearance of E.coli under a microscope following the Gram stain is pink or red
because of retaining the counter staining dye referred to as safranin. The appearance is as shown
below
3. The expected appearance of Bacillus subtilis under a light microscope following Gram stain is
rod shaped as shown below
experiments
The common sources of contamination include contamination from analysts, low quality
reagents, analysts, visitor’s entry, sampling tools, sample environment and container. To prevent
such errors, it would be ideal reagents plus quality of recommended grades from reputable
dealers. One should also maintain the recommended condition between analysis and the sample
collection. It is also important to use clean glassware during analysis and ensure care as per the
recommendations in the microbial testing labs.
Gram stain
1. The main difference between gram positive and gram negative stain is that in gram positive
cell walls have a thick layer of techoic acids referred to as peptidoglycan, while in the gram
negative bacteria, the cell walls have a thin layer without techoic acids. Besides, gram positive
bacteria do stain purple due to the thick layer mentioned above while gram negative bacteria do
stain red due to the thin peptidoglycan layer that is surrounded by the plasma membrane and
therefore will not stain with crystal violet
2. The expected appearance of E.coli under a microscope following the Gram stain is pink or red
because of retaining the counter staining dye referred to as safranin. The appearance is as shown
below
3. The expected appearance of Bacillus subtilis under a light microscope following Gram stain is
rod shaped as shown below

Streak plate and spread plate
1. The main differences between spread plate and streak plate
Streak plate technique uses solid bacterial mixture while spread plate uses both liquid and solid
mixtures. Also streak plate can be done using one plate while spread plate might require more
than one plate. For the aims, spread plate allows one to obtain one colony of each type of
bacteria from a mixture of many while streak plate achieves the same result only through a
mixture of less bacteria types
2. A procedure referred to as viable plate count was conducted. In the same context, viable
referrers to the identification of the sum of actively dividing cells in a given population.
3. The estimate of cell numbers provided by a viable plate count and the estimate based on total
cell counts give the same results because viable plate counts are known to underestimate total
number of bacteria.
Yakult
1. The meaning of “probiotic” and how it relates to Yakult.
Probiotic is a species that is believed to provide health benefits when consumed. I relates to
yakult in that it is a probiotic dairy product produced through fermenting skimmed milk with
special strain of the shirota, bacterium
2. Species found in Yakult
The bacterium found in yakult is lactobacillus casei. However, a standard Yakult contains 14
grams of sugar for every 100grams a 65ml bottle.
1. The main differences between spread plate and streak plate
Streak plate technique uses solid bacterial mixture while spread plate uses both liquid and solid
mixtures. Also streak plate can be done using one plate while spread plate might require more
than one plate. For the aims, spread plate allows one to obtain one colony of each type of
bacteria from a mixture of many while streak plate achieves the same result only through a
mixture of less bacteria types
2. A procedure referred to as viable plate count was conducted. In the same context, viable
referrers to the identification of the sum of actively dividing cells in a given population.
3. The estimate of cell numbers provided by a viable plate count and the estimate based on total
cell counts give the same results because viable plate counts are known to underestimate total
number of bacteria.
Yakult
1. The meaning of “probiotic” and how it relates to Yakult.
Probiotic is a species that is believed to provide health benefits when consumed. I relates to
yakult in that it is a probiotic dairy product produced through fermenting skimmed milk with
special strain of the shirota, bacterium
2. Species found in Yakult
The bacterium found in yakult is lactobacillus casei. However, a standard Yakult contains 14
grams of sugar for every 100grams a 65ml bottle.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

CFU ml-1
1. The number of colony forming units that are present in 100 l of every dilution include
10^-5-261 CFU/100ul
10^-6-23CFU/100ul
10^-7-1CFU/100ul
2. The number of colony forming units that were present in one ml of every dilution was
Ten times more
3. The number of cells that are present in a typical milliliter of the undiluted culture is
10/10^-7=1*10^8
4. This number relates to the advertised quantity of bacteria in a bottle of Yakult in that it is
lower compared to the quantity of bacteria in one bottle of Yakult
5. The reason as to why some colonies are not derived from the same bacterium is because
Every colony contains thousands of bacteria all from one bacterium.
Identification of bacteria – API 20E
1. The group of organisms that can be identified using API 20E test strips is the
group of bacteria like entero bacteria and lactobacilli to mention just a few.
2. The biochemical basis of tests on the API20E test strips.
The plastic strips holds about 20 mini test chambers with dehydrated media each having
chemically defined composition for every test.
They include;
OPNG test for beta galactosidfase enzyme by hydrolysis of substrate o-nitro phenyl-b-D-
galactopyranoside
ADH test for dicarboxilation of amino acids lysine by lysine decarboxylase
IND indole test for production of indole from typtophan by typtophanase enzyme
1. The number of colony forming units that are present in 100 l of every dilution include
10^-5-261 CFU/100ul
10^-6-23CFU/100ul
10^-7-1CFU/100ul
2. The number of colony forming units that were present in one ml of every dilution was
Ten times more
3. The number of cells that are present in a typical milliliter of the undiluted culture is
10/10^-7=1*10^8
4. This number relates to the advertised quantity of bacteria in a bottle of Yakult in that it is
lower compared to the quantity of bacteria in one bottle of Yakult
5. The reason as to why some colonies are not derived from the same bacterium is because
Every colony contains thousands of bacteria all from one bacterium.
Identification of bacteria – API 20E
1. The group of organisms that can be identified using API 20E test strips is the
group of bacteria like entero bacteria and lactobacilli to mention just a few.
2. The biochemical basis of tests on the API20E test strips.
The plastic strips holds about 20 mini test chambers with dehydrated media each having
chemically defined composition for every test.
They include;
OPNG test for beta galactosidfase enzyme by hydrolysis of substrate o-nitro phenyl-b-D-
galactopyranoside
ADH test for dicarboxilation of amino acids lysine by lysine decarboxylase
IND indole test for production of indole from typtophan by typtophanase enzyme
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

TDA test for detection of tryptophan deaminase enzyme.
3. How API 20E strips are incubated plus inoculated.
During API 20E incubation, metabolism gives color changes that is neither spontaneous nor
relevant by additional of reagents. However, during inoculation both the capsule and tube of the
test are filled with bacterial suspension, anaerobiosis is then created in then created and the
incubation box closed where it is incubated at 370C for about 24 hours
4. The strains of a particular species would not give the same results (profile) in an API 20E test
strip because after comparing the API 20E strips, there was no unique biochemical profile
numbers for the strains
5. Factors that change the results as seen in an API test strip is change or modification of the
procedure
Identification of bacteria – significance of species identified
1. Significance of the following bacterium in a clinical perspective
a. Serratia marcescens
The species is involved in hospitalized acquired infections especially the catheter relate
bacteremia wound and urinary tract infections
b. Proteus mirabilis
The bacterium show swarming motility as well as unrease activity
c. Pseudomonas fluorescens
The bacteria produces mupirocin antibiotic which is effective in treating different types of skin ,
eye and ear complications
d. Klebsiella pneumoniae
3. How API 20E strips are incubated plus inoculated.
During API 20E incubation, metabolism gives color changes that is neither spontaneous nor
relevant by additional of reagents. However, during inoculation both the capsule and tube of the
test are filled with bacterial suspension, anaerobiosis is then created in then created and the
incubation box closed where it is incubated at 370C for about 24 hours
4. The strains of a particular species would not give the same results (profile) in an API 20E test
strip because after comparing the API 20E strips, there was no unique biochemical profile
numbers for the strains
5. Factors that change the results as seen in an API test strip is change or modification of the
procedure
Identification of bacteria – significance of species identified
1. Significance of the following bacterium in a clinical perspective
a. Serratia marcescens
The species is involved in hospitalized acquired infections especially the catheter relate
bacteremia wound and urinary tract infections
b. Proteus mirabilis
The bacterium show swarming motility as well as unrease activity
c. Pseudomonas fluorescens
The bacteria produces mupirocin antibiotic which is effective in treating different types of skin ,
eye and ear complications
d. Klebsiella pneumoniae

Helps in identification of people with weak immune system especially the old age and also
shows a range of clinical diseases like pneumonia
e. Aeromonas veronii
The blood digested by the bacterium helps in lowering the concentration of the bacterium via the
activatons of the membrane attack complexions.
Identification of bacteria – 16S rRNA gene sequencing
1. The main advantages of 16S rRNA gene sequencing over phenotype tests
The sequencing is used in identification of microorganisms that are misidentified through
conventional means. The sequencing also provides accurate grouped organisms for further study
2. Distribution of 16 S rRNA percent identity of pairs od training set sequence being of the same
as well as different species for sdelect gram of either negative or positive gram bacteria.however,
species for which sequence variability between sequences on the same species was very high
compared to those from different species.
Importance of identification in regard to biology
Understanding how a bacteria works plus how it is structured is the same as knowing how it
affects people. For instance, if bacteria can make an individual sick and has a beta lactam ring in
its cell wall then it is ideal to understand since a drug that works on the same ring is in a position
to kill the bacteria. However, unknown viruses might have clinical uses. For instance, some
pharmaceutical medications are based on the products made by organisms. Penicillin is obtained
from fungus. Digoxin and digitalis, cardiac drugs are made from plants. Hence, there’s a
likelihood that the unidentified bacteria can have something it produces which can lead to
medical recovery
shows a range of clinical diseases like pneumonia
e. Aeromonas veronii
The blood digested by the bacterium helps in lowering the concentration of the bacterium via the
activatons of the membrane attack complexions.
Identification of bacteria – 16S rRNA gene sequencing
1. The main advantages of 16S rRNA gene sequencing over phenotype tests
The sequencing is used in identification of microorganisms that are misidentified through
conventional means. The sequencing also provides accurate grouped organisms for further study
2. Distribution of 16 S rRNA percent identity of pairs od training set sequence being of the same
as well as different species for sdelect gram of either negative or positive gram bacteria.however,
species for which sequence variability between sequences on the same species was very high
compared to those from different species.
Importance of identification in regard to biology
Understanding how a bacteria works plus how it is structured is the same as knowing how it
affects people. For instance, if bacteria can make an individual sick and has a beta lactam ring in
its cell wall then it is ideal to understand since a drug that works on the same ring is in a position
to kill the bacteria. However, unknown viruses might have clinical uses. For instance, some
pharmaceutical medications are based on the products made by organisms. Penicillin is obtained
from fungus. Digoxin and digitalis, cardiac drugs are made from plants. Hence, there’s a
likelihood that the unidentified bacteria can have something it produces which can lead to
medical recovery
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
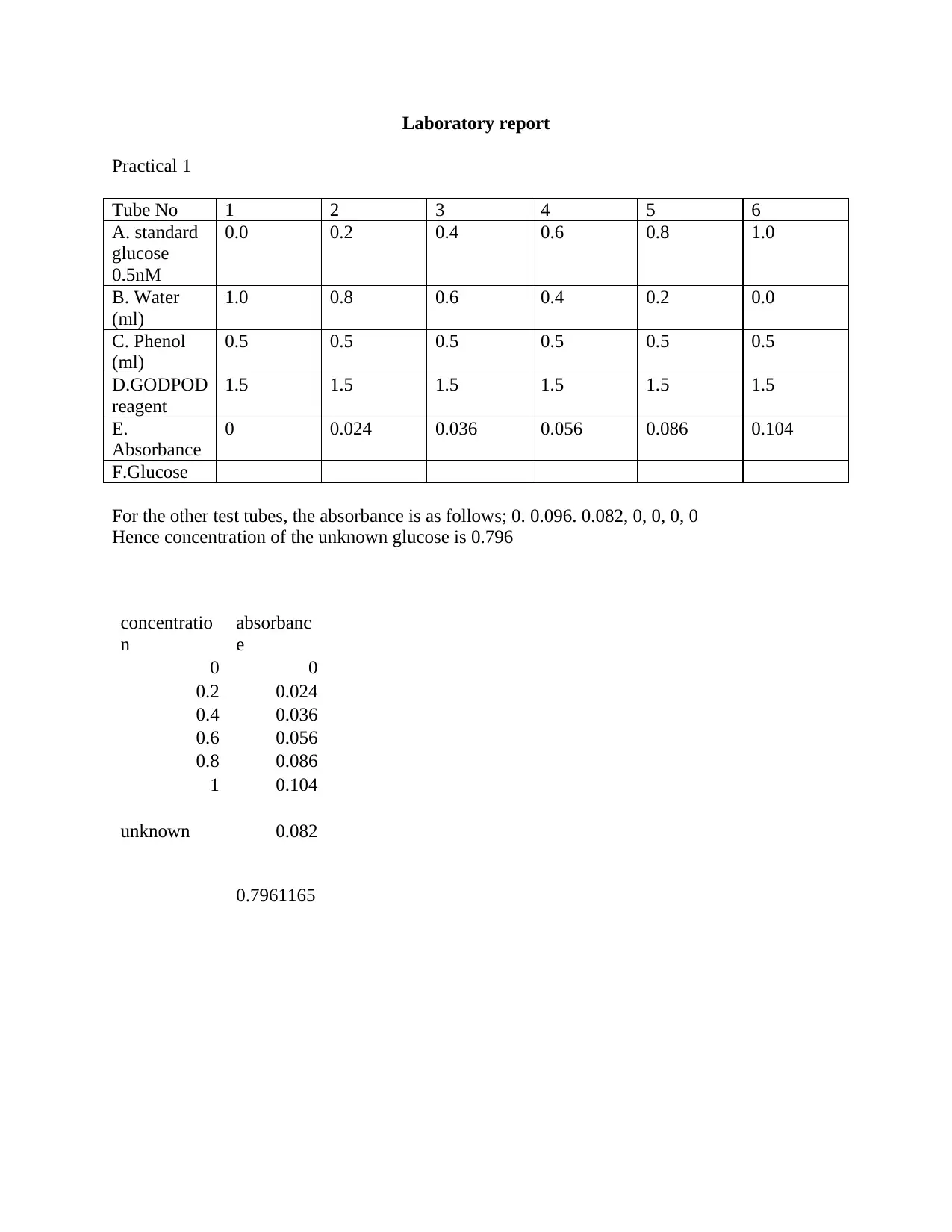
Laboratory report
Practical 1
Tube No 1 2 3 4 5 6
A. standard
glucose
0.5nM
0.0 0.2 0.4 0.6 0.8 1.0
B. Water
(ml)
1.0 0.8 0.6 0.4 0.2 0.0
C. Phenol
(ml)
0.5 0.5 0.5 0.5 0.5 0.5
D.GODPOD
reagent
1.5 1.5 1.5 1.5 1.5 1.5
E.
Absorbance
0 0.024 0.036 0.056 0.086 0.104
F.Glucose
For the other test tubes, the absorbance is as follows; 0. 0.096. 0.082, 0, 0, 0, 0
Hence concentration of the unknown glucose is 0.796
concentratio
n
absorbanc
e
0 0
0.2 0.024
0.4 0.036
0.6 0.056
0.8 0.086
1 0.104
unknown 0.082
0.7961165
Practical 1
Tube No 1 2 3 4 5 6
A. standard
glucose
0.5nM
0.0 0.2 0.4 0.6 0.8 1.0
B. Water
(ml)
1.0 0.8 0.6 0.4 0.2 0.0
C. Phenol
(ml)
0.5 0.5 0.5 0.5 0.5 0.5
D.GODPOD
reagent
1.5 1.5 1.5 1.5 1.5 1.5
E.
Absorbance
0 0.024 0.036 0.056 0.086 0.104
F.Glucose
For the other test tubes, the absorbance is as follows; 0. 0.096. 0.082, 0, 0, 0, 0
Hence concentration of the unknown glucose is 0.796
concentratio
n
absorbanc
e
0 0
0.2 0.024
0.4 0.036
0.6 0.056
0.8 0.086
1 0.104
unknown 0.082
0.7961165
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
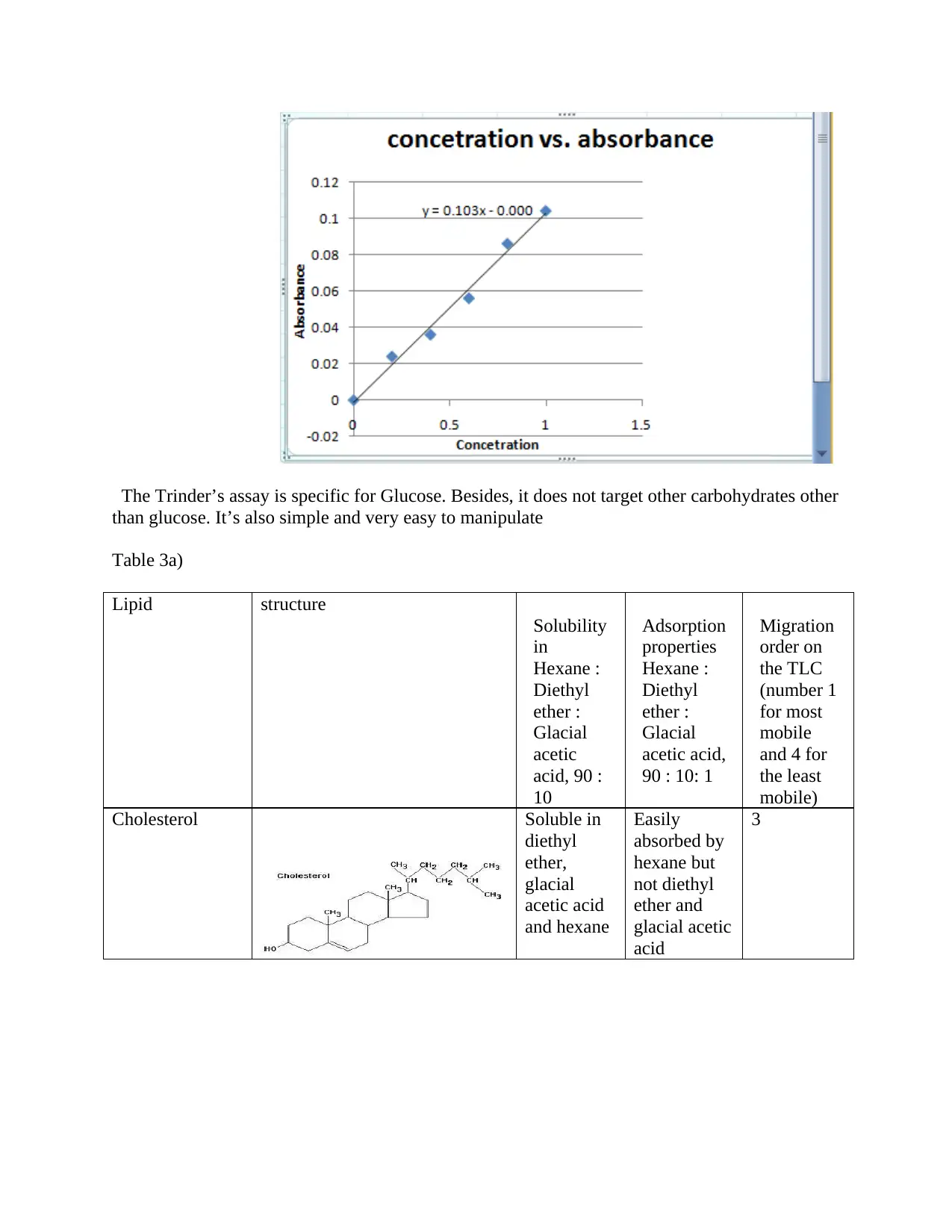
The Trinder’s assay is specific for Glucose. Besides, it does not target other carbohydrates other
than glucose. It’s also simple and very easy to manipulate
Table 3a)
Lipid structure
Solubility
in
Hexane :
Diethyl
ether :
Glacial
acetic
acid, 90 :
10
Adsorption
properties
Hexane :
Diethyl
ether :
Glacial
acetic acid,
90 : 10: 1
Migration
order on
the TLC
(number 1
for most
mobile
and 4 for
the least
mobile)
Cholesterol Soluble in
diethyl
ether,
glacial
acetic acid
and hexane
Easily
absorbed by
hexane but
not diethyl
ether and
glacial acetic
acid
3
than glucose. It’s also simple and very easy to manipulate
Table 3a)
Lipid structure
Solubility
in
Hexane :
Diethyl
ether :
Glacial
acetic
acid, 90 :
10
Adsorption
properties
Hexane :
Diethyl
ether :
Glacial
acetic acid,
90 : 10: 1
Migration
order on
the TLC
(number 1
for most
mobile
and 4 for
the least
mobile)
Cholesterol Soluble in
diethyl
ether,
glacial
acetic acid
and hexane
Easily
absorbed by
hexane but
not diethyl
ether and
glacial acetic
acid
3
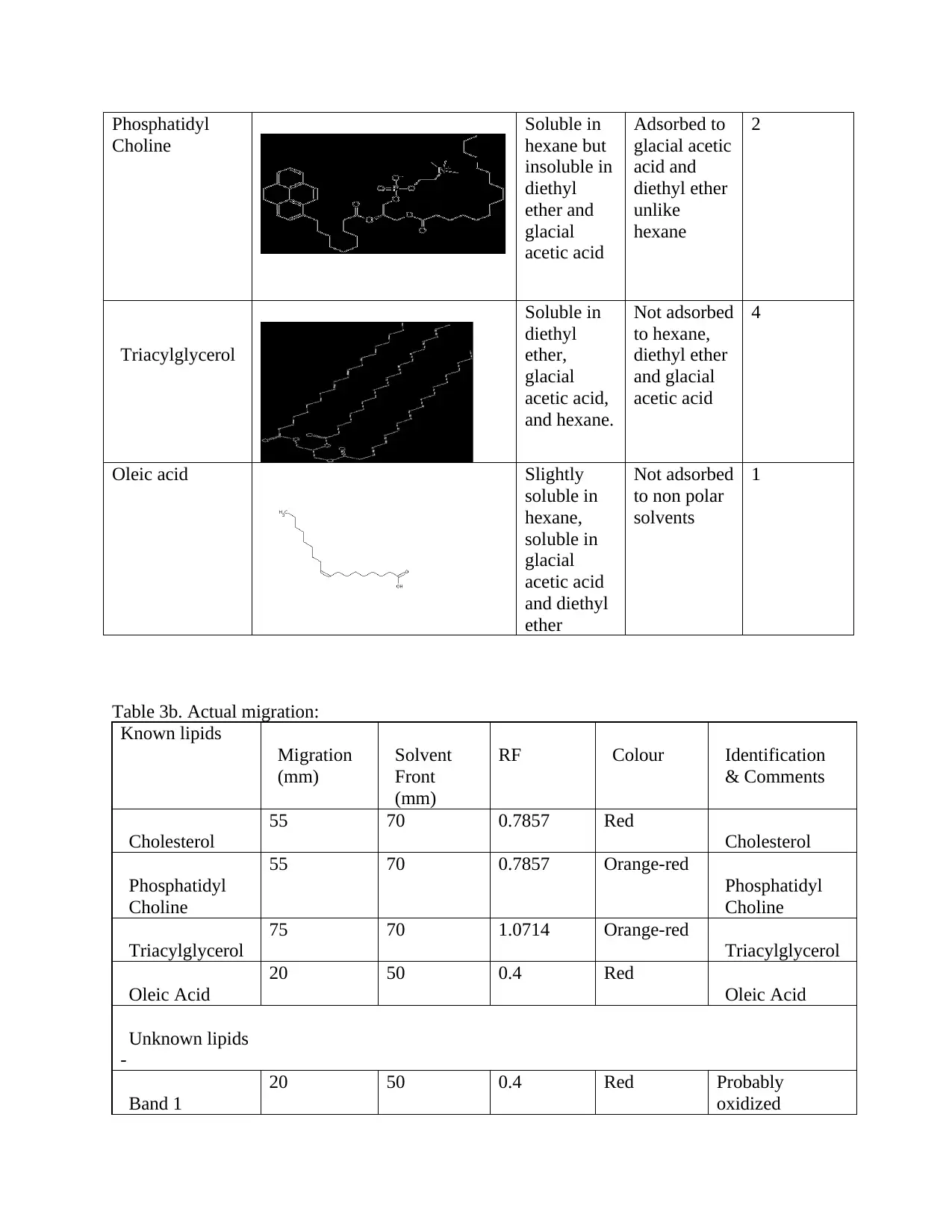
Phosphatidyl
Choline
Soluble in
hexane but
insoluble in
diethyl
ether and
glacial
acetic acid
Adsorbed to
glacial acetic
acid and
diethyl ether
unlike
hexane
2
Triacylglycerol
Soluble in
diethyl
ether,
glacial
acetic acid,
and hexane.
Not adsorbed
to hexane,
diethyl ether
and glacial
acetic acid
4
Oleic acid Slightly
soluble in
hexane,
soluble in
glacial
acetic acid
and diethyl
ether
Not adsorbed
to non polar
solvents
1
Table 3b. Actual migration:
Known lipids
Migration
(mm)
Solvent
Front
(mm)
RF Colour Identification
& Comments
Cholesterol
55 70 0.7857 Red
Cholesterol
Phosphatidyl
Choline
55 70 0.7857 Orange-red
Phosphatidyl
Choline
Triacylglycerol
75 70 1.0714 Orange-red
Triacylglycerol
Oleic Acid
20 50 0.4 Red
Oleic Acid
Unknown lipids
-
Band 1
20 50 0.4 Red Probably
oxidized
Choline
Soluble in
hexane but
insoluble in
diethyl
ether and
glacial
acetic acid
Adsorbed to
glacial acetic
acid and
diethyl ether
unlike
hexane
2
Triacylglycerol
Soluble in
diethyl
ether,
glacial
acetic acid,
and hexane.
Not adsorbed
to hexane,
diethyl ether
and glacial
acetic acid
4
Oleic acid Slightly
soluble in
hexane,
soluble in
glacial
acetic acid
and diethyl
ether
Not adsorbed
to non polar
solvents
1
Table 3b. Actual migration:
Known lipids
Migration
(mm)
Solvent
Front
(mm)
RF Colour Identification
& Comments
Cholesterol
55 70 0.7857 Red
Cholesterol
Phosphatidyl
Choline
55 70 0.7857 Orange-red
Phosphatidyl
Choline
Triacylglycerol
75 70 1.0714 Orange-red
Triacylglycerol
Oleic Acid
20 50 0.4 Red
Oleic Acid
Unknown lipids
-
Band 1
20 50 0.4 Red Probably
oxidized
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 18
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2025 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.
