Understanding DNA Analysis and Molecular Biology Techniques
VerifiedAdded on 2020/05/16
|8
|1930
|161
AI Summary
The assignment provides a comprehensive overview of DNA's structural dynamics, including the helical models proposed by Watson and Crick, as well as alternative structures like A-Form, B-Form, and Z-Form. It examines the replication process with a focus on the polymerase chain reaction (PCR), detailing its steps such as denaturation, annealing, and extension. Additionally, it explores Chromatin Immunoprecipitation Sequencing (ChIP) for mapping protein-DNA interactions, crucial in understanding gene regulation mechanisms. The assignment also highlights applications of these molecular techniques in identifying transcription factors' roles in phenotypic expressions and genetic research advancements.

UNIVERSITY:
NAME :
STUDENT ID:
COURSE CODE
COURSE NAME
Molecular Biology
NAME :
STUDENT ID:
COURSE CODE
COURSE NAME
Molecular Biology
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Molecular Biology 2
HOME WORK 2
Question 1
The double stranding of DNA can be made to be reversible and denatured at various
melting points. The transition from double stranded formation helix to single strand helix,
changes the absorbance of the bases at λ=260 which is associated with the transition phase.
Absorbance at A260, can increase when the DNA melts reaching hyper chromatic effect.
The thermal melting point of any DNA has shown that as the temperature is increased,
the solution of double strand bonds denatures, leading to formation of single strand
structures. The temperature level which the DNA melts is referred to as TM. The melting
levels is often detected by the absorbance of 260 nm, while the coefficient of extinction in the
double strand DNA is lowered to approximately 30% which is lower than the single strand
DNA. Lowered absorption at 260 nm in the double strand DNA is often occasioned by the
interactions of the dipoles present on the stacked bases, which is interrupted by the DNA
single stranding.
Responsible forces which influence this effect include:
Hydrogen bonds
- Have strong stability on the double strand DNA stability. However as the TM of the
double strand increases the GC content also rises. This leads to effect on the differences on
the stacking interactions and the presence of hydrogen bonds on the GC players which play a
role affecting the melting of DNA.
Electrostatic forces
- The charged repulsion of the phosphate compounds in the DNA leads to
destabilization of the bases, and it has been seen as much stronger than the non polar chains
present in proteins. The bases which shows strong attachment with GC pair and the AT bases
have shown to increase in TM as the content of GC increases
Van der Waals forces
- They have a significant stability on the double strands of DNA. They are
characterised by strong dipole interactions amongst the bases and stronger on the non polar
chains of protein. The bases which are stronger stacking with GC base pairs and AT base
pairs have shown increase the overall TM as the GC content increases.
Question 2
The stability of proteins has been found to be associated with direct associations
between atoms on its backbone chain. Also increase on hydrophobic on the amino side chain
are exposed. Hydrophobic does not really signify any formal interactions but rather the ability
HOME WORK 2
Question 1
The double stranding of DNA can be made to be reversible and denatured at various
melting points. The transition from double stranded formation helix to single strand helix,
changes the absorbance of the bases at λ=260 which is associated with the transition phase.
Absorbance at A260, can increase when the DNA melts reaching hyper chromatic effect.
The thermal melting point of any DNA has shown that as the temperature is increased,
the solution of double strand bonds denatures, leading to formation of single strand
structures. The temperature level which the DNA melts is referred to as TM. The melting
levels is often detected by the absorbance of 260 nm, while the coefficient of extinction in the
double strand DNA is lowered to approximately 30% which is lower than the single strand
DNA. Lowered absorption at 260 nm in the double strand DNA is often occasioned by the
interactions of the dipoles present on the stacked bases, which is interrupted by the DNA
single stranding.
Responsible forces which influence this effect include:
Hydrogen bonds
- Have strong stability on the double strand DNA stability. However as the TM of the
double strand increases the GC content also rises. This leads to effect on the differences on
the stacking interactions and the presence of hydrogen bonds on the GC players which play a
role affecting the melting of DNA.
Electrostatic forces
- The charged repulsion of the phosphate compounds in the DNA leads to
destabilization of the bases, and it has been seen as much stronger than the non polar chains
present in proteins. The bases which shows strong attachment with GC pair and the AT bases
have shown to increase in TM as the content of GC increases
Van der Waals forces
- They have a significant stability on the double strands of DNA. They are
characterised by strong dipole interactions amongst the bases and stronger on the non polar
chains of protein. The bases which are stronger stacking with GC base pairs and AT base
pairs have shown increase the overall TM as the GC content increases.
Question 2
The stability of proteins has been found to be associated with direct associations
between atoms on its backbone chain. Also increase on hydrophobic on the amino side chain
are exposed. Hydrophobic does not really signify any formal interactions but rather the ability
Molecular Biology 3
of no polar residues to affect the overall structure of the and the fluctuation of aqueous
solvent.
The observable manifestation is the increase in the heat produced on the overall system,
ΔCp, which results in strong temperature remittance on the transition process of enthalpy and
entropy. Smalls deviations from the temperature results in thermal contribution which results
in ΔH and TΔS neutralizing each other, when summed they give free energy ΔG..
But with bigger temperature range, the different functional groups of the enthalpy and
entropy files high contribution of heat on the transition seen I the free energy expulsion. At
times it has been observed to dominate the stability of the proteins and the ability to lead to
cold denaturization process.
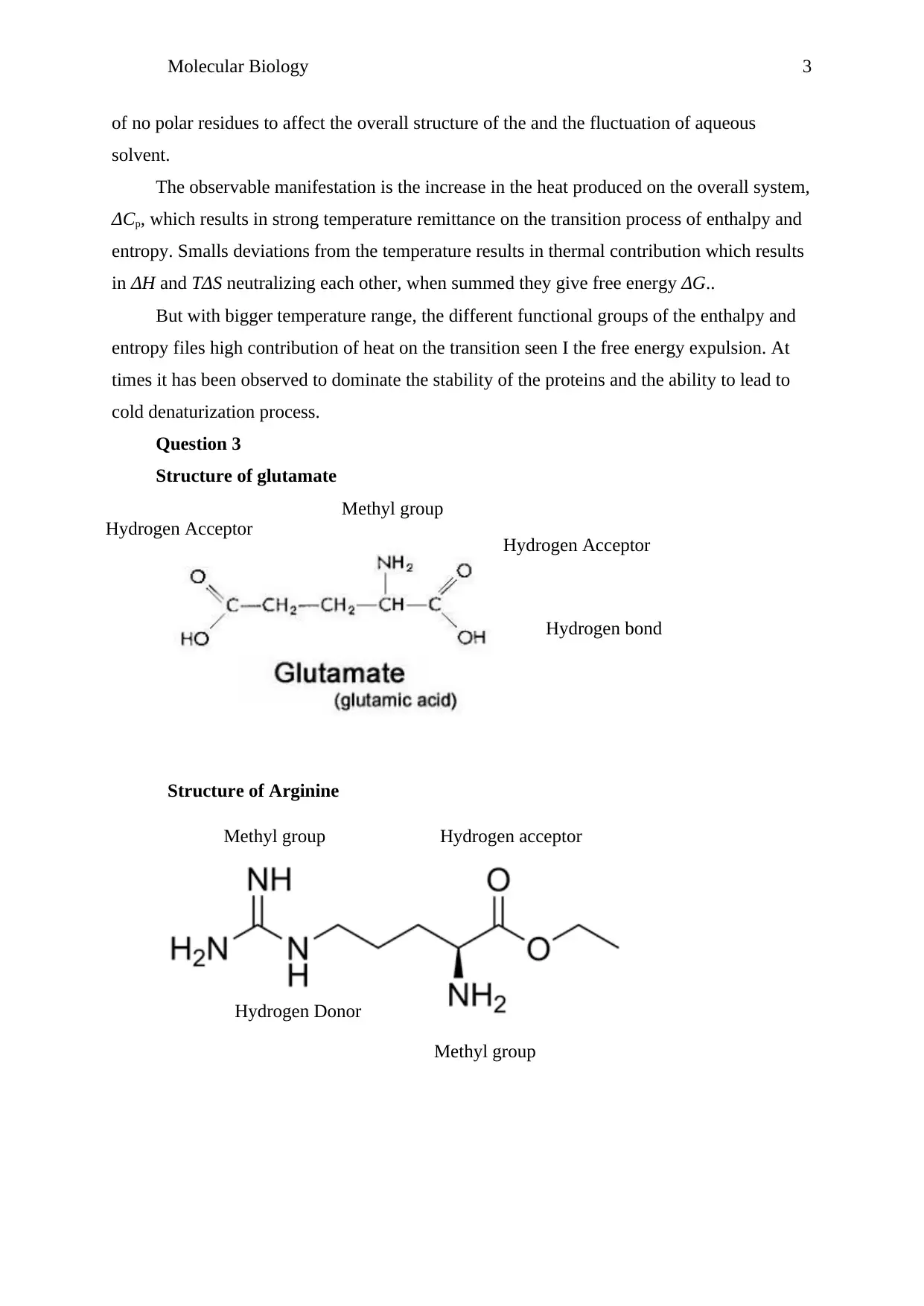
Question 3
Structure of glutamate
Structure of Arginine
Methyl group
Hydrogen bond
Hydrogen Acceptor
Hydrogen Acceptor
Hydrogen acceptor
Methyl group
Methyl group
Hydrogen Donor
of no polar residues to affect the overall structure of the and the fluctuation of aqueous
solvent.
The observable manifestation is the increase in the heat produced on the overall system,
ΔCp, which results in strong temperature remittance on the transition process of enthalpy and
entropy. Smalls deviations from the temperature results in thermal contribution which results
in ΔH and TΔS neutralizing each other, when summed they give free energy ΔG..
But with bigger temperature range, the different functional groups of the enthalpy and
entropy files high contribution of heat on the transition seen I the free energy expulsion. At
times it has been observed to dominate the stability of the proteins and the ability to lead to
cold denaturization process.
Question 3
Structure of glutamate
Structure of Arginine
Methyl group
Hydrogen bond
Hydrogen Acceptor
Hydrogen Acceptor
Hydrogen acceptor
Methyl group
Methyl group
Hydrogen Donor
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
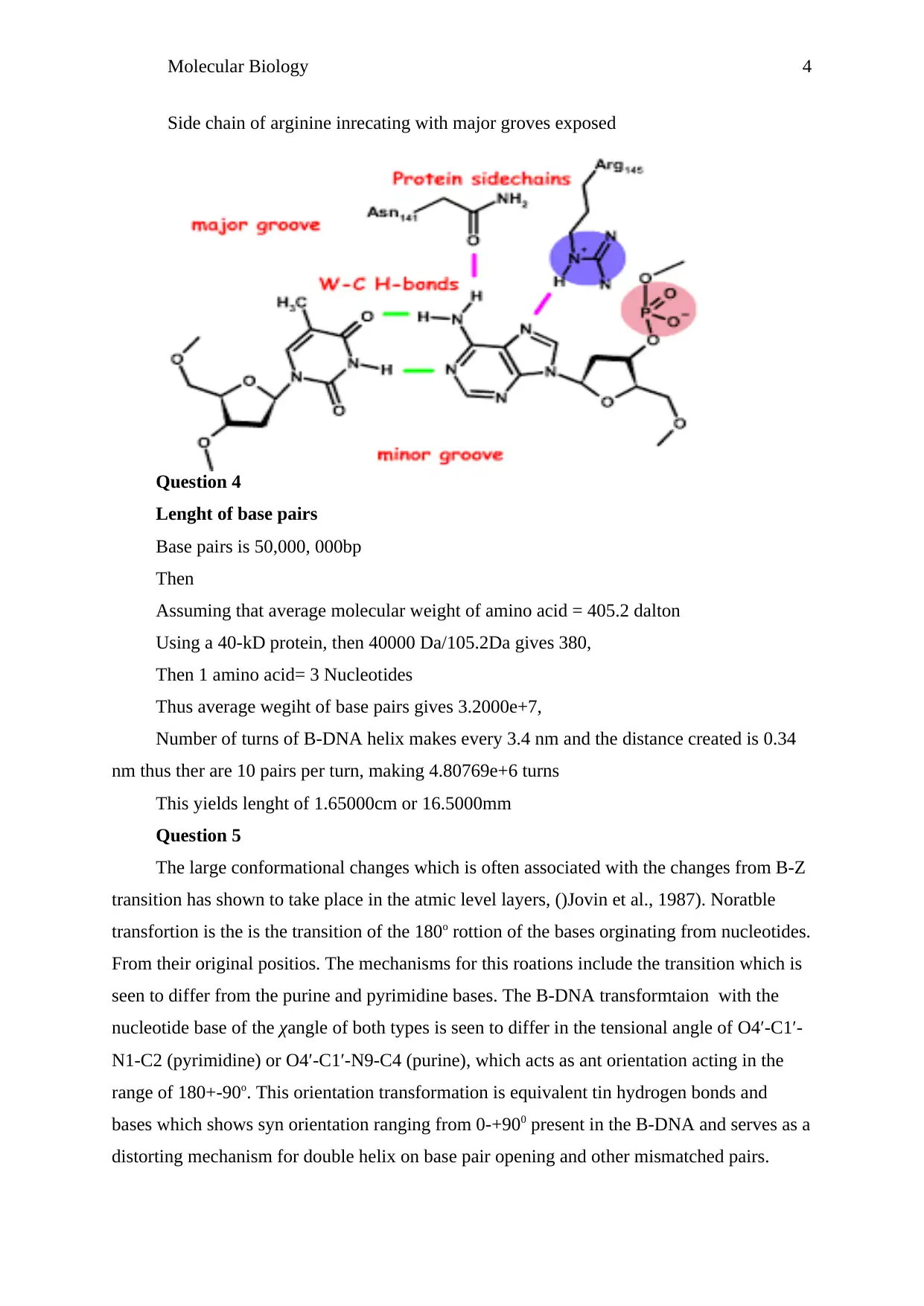
Molecular Biology 4
Side chain of arginine inrecating with major groves exposed
Question 4
Lenght of base pairs
Base pairs is 50,000, 000bp
Then
Assuming that average molecular weight of amino acid = 405.2 dalton
Using a 40-kD protein, then 40000 Da/105.2Da gives 380,
Then 1 amino acid= 3 Nucleotides
Thus average wegiht of base pairs gives 3.2000e+7,
Number of turns of B-DNA helix makes every 3.4 nm and the distance created is 0.34
nm thus ther are 10 pairs per turn, making 4.80769e+6 turns
This yields lenght of 1.65000cm or 16.5000mm
Question 5
The large conformational changes which is often associated with the changes from B-Z
transition has shown to take place in the atmic level layers, ()Jovin et al., 1987). Noratble
transfortion is the is the transition of the 180o rottion of the bases orginating from nucleotides.
From their original positios. The mechanisms for this roations include the transition which is
seen to differ from the purine and pyrimidine bases. The B-DNA transformtaion with the
nucleotide base of the χangle of both types is seen to differ in the tensional angle of O4′-C1′-
N1-C2 (pyrimidine) or O4′-C1′-N9-C4 (purine), which acts as ant orientation acting in the
range of 180+-90o. This orientation transformation is equivalent tin hydrogen bonds and
bases which shows syn orientation ranging from 0-+900 present in the B-DNA and serves as a
distorting mechanism for double helix on base pair opening and other mismatched pairs.
Side chain of arginine inrecating with major groves exposed
Question 4
Lenght of base pairs
Base pairs is 50,000, 000bp
Then
Assuming that average molecular weight of amino acid = 405.2 dalton
Using a 40-kD protein, then 40000 Da/105.2Da gives 380,
Then 1 amino acid= 3 Nucleotides
Thus average wegiht of base pairs gives 3.2000e+7,
Number of turns of B-DNA helix makes every 3.4 nm and the distance created is 0.34
nm thus ther are 10 pairs per turn, making 4.80769e+6 turns
This yields lenght of 1.65000cm or 16.5000mm
Question 5
The large conformational changes which is often associated with the changes from B-Z
transition has shown to take place in the atmic level layers, ()Jovin et al., 1987). Noratble
transfortion is the is the transition of the 180o rottion of the bases orginating from nucleotides.
From their original positios. The mechanisms for this roations include the transition which is
seen to differ from the purine and pyrimidine bases. The B-DNA transformtaion with the
nucleotide base of the χangle of both types is seen to differ in the tensional angle of O4′-C1′-
N1-C2 (pyrimidine) or O4′-C1′-N9-C4 (purine), which acts as ant orientation acting in the
range of 180+-90o. This orientation transformation is equivalent tin hydrogen bonds and
bases which shows syn orientation ranging from 0-+900 present in the B-DNA and serves as a
distorting mechanism for double helix on base pair opening and other mismatched pairs.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Molecular Biology 5
HOME WORK 3
Question 1
Fragments order
Order ranging from the smllaest to largest
BstAPI 179
Bgl 245
Bceai 387
Thl 781
Drill 908
BceAl 1292
Bsal 1779
Bpml 1784
Begl 2215
Sspl 2501
Eco01091 2674
Question 2
a) Labelling of 3’ and 5’ cuts by BamHi with the encodiing BamH I ....... 5'-
GGATCC-3'. The enzyme identifies the sequence and leaves the strand structure appearing
longer than the other. The end of these looks like this
……..G-3’
…..CCTACG-5’
HOME WORK 3
Question 1
Fragments order
Order ranging from the smllaest to largest
BstAPI 179
Bgl 245
Bceai 387
Thl 781
Drill 908
BceAl 1292
Bsal 1779
Bpml 1784
Begl 2215
Sspl 2501
Eco01091 2674
Question 2
a) Labelling of 3’ and 5’ cuts by BamHi with the encodiing BamH I ....... 5'-
GGATCC-3'. The enzyme identifies the sequence and leaves the strand structure appearing
longer than the other. The end of these looks like this
……..G-3’
…..CCTACG-5’

Molecular Biology 6
b) Hydroxyl and Phosphoryl groups
The hydroxyl groups are ploar and often not charged at physiological pH. The hydroxyl
normally ionizes at ptattisoum potentil of 10 to yiled phenolate anion, thus they are highly
inonizing. Hydroxyl group will; attached iteself on 3’ as it attaches itself on Cytsoine amino
acid.
Phosphryl groups are derived from phosphoric acid. When the phospahet compund is
linked to carbon it atahces itself to carbon atom and it is reffered to as phospahet ester. On the
compudns above, it attches itself on 5’ end.
Question 3
Micro array is a lab tool which is useful in detecting the expression of genes at the same
period in time. DNA micro array are ussuallly microspic slides which are covered by printed
aspects of tiny spots indefined positions which each spot has DNA sequcen or gene. The
DNA attahcmen to each other shows gene expressions or set of messanger of RNA expressed
through genes groups.
Northern blotting refers entails transfer of RNA to memberane , it is separted according
to size under probing technique. Hybridization process often takes place in northern blotting
through agarose gel memebrane.
Western blotting have shown similar display on the array blot as observed with northen
blotinng. The western blotting utlizes the same theoritical appraoch as seen in northern blotts.
Question 4
Polymerase chain reaction is a replication method for DNA. It makes various copies of
any segment of DNA. It can be able to take sevral amounts of DNA in single molecule and
provide amplification, (Flanagan et al., 2014).
The basics of PCR Cycling include various steps in cycling reactiopns which is often
repated in 20 to 40 cylces. It is done in automated theremo cycler and has the ability to heat
and cool reactinons , (MagdMateal, 2015).
PCR involvces various steps which include, denaturation, annealing and extension.
Denaturization takes placve at 95 degrees in 30 secosn while annealing takes place at 55-60
degrees celcius in 30 sec and finaly the extention takes place at 72 degrees and time depnds
on the size of the produt.
Denaturization stage takes place at 94 degrees celicus whereby the double strand melsts
and opens up to form single strand DNA, at this stage alll enzymatic reactions stops. At
annealing stage, the hydrogen bonds are continuously formed and borken in between the
single strands primer, (Verm et al., 2013)
b) Hydroxyl and Phosphoryl groups
The hydroxyl groups are ploar and often not charged at physiological pH. The hydroxyl
normally ionizes at ptattisoum potentil of 10 to yiled phenolate anion, thus they are highly
inonizing. Hydroxyl group will; attached iteself on 3’ as it attaches itself on Cytsoine amino
acid.
Phosphryl groups are derived from phosphoric acid. When the phospahet compund is
linked to carbon it atahces itself to carbon atom and it is reffered to as phospahet ester. On the
compudns above, it attches itself on 5’ end.
Question 3
Micro array is a lab tool which is useful in detecting the expression of genes at the same
period in time. DNA micro array are ussuallly microspic slides which are covered by printed
aspects of tiny spots indefined positions which each spot has DNA sequcen or gene. The
DNA attahcmen to each other shows gene expressions or set of messanger of RNA expressed
through genes groups.
Northern blotting refers entails transfer of RNA to memberane , it is separted according
to size under probing technique. Hybridization process often takes place in northern blotting
through agarose gel memebrane.
Western blotting have shown similar display on the array blot as observed with northen
blotinng. The western blotting utlizes the same theoritical appraoch as seen in northern blotts.
Question 4
Polymerase chain reaction is a replication method for DNA. It makes various copies of
any segment of DNA. It can be able to take sevral amounts of DNA in single molecule and
provide amplification, (Flanagan et al., 2014).
The basics of PCR Cycling include various steps in cycling reactiopns which is often
repated in 20 to 40 cylces. It is done in automated theremo cycler and has the ability to heat
and cool reactinons , (MagdMateal, 2015).
PCR involvces various steps which include, denaturation, annealing and extension.
Denaturization takes placve at 95 degrees in 30 secosn while annealing takes place at 55-60
degrees celcius in 30 sec and finaly the extention takes place at 72 degrees and time depnds
on the size of the produt.
Denaturization stage takes place at 94 degrees celicus whereby the double strand melsts
and opens up to form single strand DNA, at this stage alll enzymatic reactions stops. At
annealing stage, the hydrogen bonds are continuously formed and borken in between the
single strands primer, (Verm et al., 2013)
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Molecular Biology 7
Extension process takes place at 72 degrees celcius and is based on process and uses
DNA TO replicate. The bases are charcatrised by 3 inch side,which the polymerase adds the
dNTP from 5 to the 3 side which forms complimentary templates, (Ahangari et al., 2015)
Question 5
DNA finger printing is a method of finger printing which investigates the specificty of
DNA binding proteins invitro. It analyses protein DNA from outside and inside ofcells. DNA
foot printing techniques are essential in esnuring that proteins binds in regions of DNA .
Polymerase chain reaction magnifies the region of concern which have the potentila protein
binding sites. Protein of interets is added on the part labeelled protein, then clevage agen is
added as achemical enzyme. Proteins wil specifcally bidn to the site to the DNA template
whcih protect the DNA. When this smaples are run in polyaccrylmide polymerase, the DBA
portion template not having protein will be cut out. The template will result in distribution
with breaks in it illustrating foot prints left.
On the other hand ChIP sequcning analses protein sequencing in DNA. It combines
with chromatic immunoprecipitation which has massive DNA sequencing to identy the
binding sites in DNAlinked to prteins. It can be utilised in mapping global mappiong sites in
anylsying any protein of interets.
I is primarily used in detremining how transcripttion factos inlfuence phenotype
mechanimss. It is important in detremining how proteins interact with proteins in regulating
gene expression and understanding bilogical process involved.
ChIP selcetively enriches DNA sequences on a partilar protin in living cells. It enriches
the crosslinks of DNA in protein complexes whcih uses antibody agianst the protein of
interest. Oligonucleotide are beneficial in this process as they are added to strecthes of DNA
bound to parallele sequencing.
The chIP DNA fragments are sequencd ran and scanning process undertaken compared
to foot priiting, it requires large sets of tilling arrays which providses lower resolltuion focus.
Extension process takes place at 72 degrees celcius and is based on process and uses
DNA TO replicate. The bases are charcatrised by 3 inch side,which the polymerase adds the
dNTP from 5 to the 3 side which forms complimentary templates, (Ahangari et al., 2015)
Question 5
DNA finger printing is a method of finger printing which investigates the specificty of
DNA binding proteins invitro. It analyses protein DNA from outside and inside ofcells. DNA
foot printing techniques are essential in esnuring that proteins binds in regions of DNA .
Polymerase chain reaction magnifies the region of concern which have the potentila protein
binding sites. Protein of interets is added on the part labeelled protein, then clevage agen is
added as achemical enzyme. Proteins wil specifcally bidn to the site to the DNA template
whcih protect the DNA. When this smaples are run in polyaccrylmide polymerase, the DBA
portion template not having protein will be cut out. The template will result in distribution
with breaks in it illustrating foot prints left.
On the other hand ChIP sequcning analses protein sequencing in DNA. It combines
with chromatic immunoprecipitation which has massive DNA sequencing to identy the
binding sites in DNAlinked to prteins. It can be utilised in mapping global mappiong sites in
anylsying any protein of interets.
I is primarily used in detremining how transcripttion factos inlfuence phenotype
mechanimss. It is important in detremining how proteins interact with proteins in regulating
gene expression and understanding bilogical process involved.
ChIP selcetively enriches DNA sequences on a partilar protin in living cells. It enriches
the crosslinks of DNA in protein complexes whcih uses antibody agianst the protein of
interest. Oligonucleotide are beneficial in this process as they are added to strecthes of DNA
bound to parallele sequencing.
The chIP DNA fragments are sequencd ran and scanning process undertaken compared
to foot priiting, it requires large sets of tilling arrays which providses lower resolltuion focus.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Molecular Biology 8
References
Ahangari, G., Pornour, M., Aminzadeh, S., Bakhtou, H., & Ahmadkhaniha, H. R. (2015).
Significant Association between Catechol Amine O-Methyl Transferase (COMT) Gene
Expression Changes and Breast Cancer Pathogenesis. J Carcinog Mutagen, 6(219), 2.
El-Magd, M. A., Zaki, A. G., Kahilo, K. A., Barakat, M. E. S., & Hassan, I. F. (2014). Effect
of SNPs in Prolactin Promoter on Milk Traits in Egyptian Buffalo. Advances in Dairy
Research, 1-4.
Flanagan PV et al. Dynamic Changes in Aromatic Hydrocarbon Associated Catabolic Gene
Profiles Linked to Aerobic and Anaerobic Microcosm Studies. Innovative Energy
Policies. 2014; 3:111.
Jovin, T. M., Soumpasis, D. M., & McIntosh, L. P. (1987). The transition between b-DNA
and z-DNA. Annual Review of Physical Chemistry, 38(1), 521-558.
Verma, S., Kumar, M., Kashyap, S., Singh, M., & Venkatesh, V. (2013). Current senario of
Escherichia coli and its serotype ‘‘O157: H7’’in Indian subcontinent. IJIRSET, 2, 2641-
2644.
References
Ahangari, G., Pornour, M., Aminzadeh, S., Bakhtou, H., & Ahmadkhaniha, H. R. (2015).
Significant Association between Catechol Amine O-Methyl Transferase (COMT) Gene
Expression Changes and Breast Cancer Pathogenesis. J Carcinog Mutagen, 6(219), 2.
El-Magd, M. A., Zaki, A. G., Kahilo, K. A., Barakat, M. E. S., & Hassan, I. F. (2014). Effect
of SNPs in Prolactin Promoter on Milk Traits in Egyptian Buffalo. Advances in Dairy
Research, 1-4.
Flanagan PV et al. Dynamic Changes in Aromatic Hydrocarbon Associated Catabolic Gene
Profiles Linked to Aerobic and Anaerobic Microcosm Studies. Innovative Energy
Policies. 2014; 3:111.
Jovin, T. M., Soumpasis, D. M., & McIntosh, L. P. (1987). The transition between b-DNA
and z-DNA. Annual Review of Physical Chemistry, 38(1), 521-558.
Verma, S., Kumar, M., Kashyap, S., Singh, M., & Venkatesh, V. (2013). Current senario of
Escherichia coli and its serotype ‘‘O157: H7’’in Indian subcontinent. IJIRSET, 2, 2641-
2644.
1 out of 8
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2025 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.