Evaluating Pathogenic Bacteria in Commercial Raw Meat Dog Food
VerifiedAdded on 2023/01/17
|5
|1337
|56
Practical Assignment
AI Summary
This practical assignment outlines a detailed methodology for detecting and analyzing pathogenic bacteria in commercially available raw meat dog food. The experiment involves testing eight samples from two different brands (Natures Menu and Natural Instincts) using selective agar plates, including Listeria Brilliance agar, Salmonella chromogenic agar, Brilliance E. coli agar, and Plate count agar. The procedure includes preparing agar plates, enriching food samples in appropriate broths, inoculating the broths onto selective agars, and incubating them under specific conditions. Positive and negative controls are used to validate the results. The assignment also describes how to calculate Colony Forming Units (CFU) and bacterial growth rates, providing a comprehensive approach to assessing the microbiological safety of raw meat dog food. Desklib offers a platform for students to access this and similar solved assignments.

“Presence of pathogenic bacteria in commercially available raw meat dog food”
SAMPLES:
No of samples: 4+4=8
Natures Menu
• poultry hearts
• Beef chunks
• chicken
• lamb
Natural instincts
• pure chicken
• Pure Beef
• Natural Lamb
• Pork
No of Brands: 2 (Natures menu, Natural instincts)
No of sampling=3
SELECTIVE AGAR PLATES:
Listeria Brilliance- for isolation, enumeration and presumptive
identification of Listeria species and Listeria monocytogenes from food
samples
Salmonella chromogenic- for the identification of Salmonella species from
other organisms in the family Enterobacteriaceae.
Brilliance E. coli agar- for the detection and enumeration of Escherichia
coli and other coliforms from food and water samples.
Plate count agar- for the enumeration of viable organisms in food, animal
feed and water samples
Table 1: Controls
Sample Negative controls Positive controls
Listeria Brilliance Enterococcus feacalis Listeria monocytogenes
Listeria innocua
Salmonella
chromogenic
Escherichia coli
Pseudomonas
aeruginosa
Salmonella enteritidis
Salmonella poona
Brilliance E. coli agar Staphylococcus aureus Escherichia coli
Klebsiella pneumoniae
Plate count agar Uninoculated medium Escherichia coli
Bacillus subtilis
Staphylococcus aureus
SAMPLES:
No of samples: 4+4=8
Natures Menu
• poultry hearts
• Beef chunks
• chicken
• lamb
Natural instincts
• pure chicken
• Pure Beef
• Natural Lamb
• Pork
No of Brands: 2 (Natures menu, Natural instincts)
No of sampling=3
SELECTIVE AGAR PLATES:
Listeria Brilliance- for isolation, enumeration and presumptive
identification of Listeria species and Listeria monocytogenes from food
samples
Salmonella chromogenic- for the identification of Salmonella species from
other organisms in the family Enterobacteriaceae.
Brilliance E. coli agar- for the detection and enumeration of Escherichia
coli and other coliforms from food and water samples.
Plate count agar- for the enumeration of viable organisms in food, animal
feed and water samples
Table 1: Controls
Sample Negative controls Positive controls
Listeria Brilliance Enterococcus feacalis Listeria monocytogenes
Listeria innocua
Salmonella
chromogenic
Escherichia coli
Pseudomonas
aeruginosa
Salmonella enteritidis
Salmonella poona
Brilliance E. coli agar Staphylococcus aureus Escherichia coli
Klebsiella pneumoniae
Plate count agar Uninoculated medium Escherichia coli
Bacillus subtilis
Staphylococcus aureus
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

1) Prepare 60 agar plates for each of the selective media a day before
the actual experiment for determination of pathogenic bacteria in the
food samples
For 8 samples/per agar media=20
8 sample in duplicate (8*2=16) + positive controls duplicate (1*2=2) + Negative
controls duplicate (1*2=2)
No of sampling (3) = 20*3=60
2) Prepare 80 (8*10) agar plates of Plate count agar for calculation of
COLONY FORMING UNITS (CFU) and GROWTH RATE
Determination of pathogenic bacteria in the food samples
A) LISTERIA in Listeria Brilliance agar 1
1. Add 25g of food sample to 225ml of ONE Broth-Listeria (CM1066 &
SR0234) and stomach for a minimum of 30 seconds to mix the sample on
day1.
2. Incubate the broth at 30ºC for 24 ± 2 hours.
3. On day 2 Gently agitate the bag then, using a microbiological loop,
remove 10μl and inoculate onto an Brilliance Listeria Agar plate and
incubate at 37ºC for 24 ± 2 hours. Examine the plate for blue colonies
with and without opaque white halos.
4. Confirm presumptive colonies on the agar plate as Listeria
monocytogenes or Listeria species by appropriate methods e.g. Gram
stain, catalase, Oxoid O.B.I.S. mono ID0600M, Oxoid Listeria Latex Test Kit
DR1126A, Microbact Listeria 12L MB1128A.
Positive and negative controls
1. Streak a plate of nutrient agar with a glycerol stock of organisms listed as
positive and negative controls in Table-1 on Day 0.
2. On day 1 add one colony to 100 ml of nutrient broth and incubate the broth at
30ºC for 24 ± 2 hours.
3. On day 2 Gently agitate the culture, using a microbiological loop, remove 10μl
and inoculate onto an Brilliance Listeria Agar plate and incubate at 37ºC for 24 ±
2 hours. Examine the plate for blue colonies with and without opaque white
halos.
CFU/ml= (No of colonies X dilution factor)/ volume of culture on plate
B) Salmonella in Salmonella chromogenic agar plate2
the actual experiment for determination of pathogenic bacteria in the
food samples
For 8 samples/per agar media=20
8 sample in duplicate (8*2=16) + positive controls duplicate (1*2=2) + Negative
controls duplicate (1*2=2)
No of sampling (3) = 20*3=60
2) Prepare 80 (8*10) agar plates of Plate count agar for calculation of
COLONY FORMING UNITS (CFU) and GROWTH RATE
Determination of pathogenic bacteria in the food samples
A) LISTERIA in Listeria Brilliance agar 1
1. Add 25g of food sample to 225ml of ONE Broth-Listeria (CM1066 &
SR0234) and stomach for a minimum of 30 seconds to mix the sample on
day1.
2. Incubate the broth at 30ºC for 24 ± 2 hours.
3. On day 2 Gently agitate the bag then, using a microbiological loop,
remove 10μl and inoculate onto an Brilliance Listeria Agar plate and
incubate at 37ºC for 24 ± 2 hours. Examine the plate for blue colonies
with and without opaque white halos.
4. Confirm presumptive colonies on the agar plate as Listeria
monocytogenes or Listeria species by appropriate methods e.g. Gram
stain, catalase, Oxoid O.B.I.S. mono ID0600M, Oxoid Listeria Latex Test Kit
DR1126A, Microbact Listeria 12L MB1128A.
Positive and negative controls
1. Streak a plate of nutrient agar with a glycerol stock of organisms listed as
positive and negative controls in Table-1 on Day 0.
2. On day 1 add one colony to 100 ml of nutrient broth and incubate the broth at
30ºC for 24 ± 2 hours.
3. On day 2 Gently agitate the culture, using a microbiological loop, remove 10μl
and inoculate onto an Brilliance Listeria Agar plate and incubate at 37ºC for 24 ±
2 hours. Examine the plate for blue colonies with and without opaque white
halos.
CFU/ml= (No of colonies X dilution factor)/ volume of culture on plate
B) Salmonella in Salmonella chromogenic agar plate2

1. Add 25g of food sample to 225ml of Rappaport-Vassiliadis Enrichment
Broth (CM0669), and stomach for a minimum of 30 seconds to mix the
sample on day1.
2. Incubate the broth at 30ºC for 24 ± 2 hours.
3. On day 2 Gently agitate the bag then, using a microbiological loop,
remove 10μl and inoculate onto an Salmonella chromogenic Agar plate
and incubate at 37ºC for 24 ± 2 hours. Examine the plates for coloured
colonies.
Positive and negative controls
1. Streak a plate of nutrient agar with a glycerol stock of organisms listed as
positive and negative controls in Table-1 on Day 0.
2. On day 1 add one colony to 100 ml of nutrient broth and incubate the broth at
30ºC for 24 ± 2 hours.
3. On day 2 Gently agitate the culture, using a microbiological loop, remove 10μl
and inoculate onto an Salmonella chromogenic Agar plate and incubate at 37ºC
for 24 ± 2 hours. Examine the plates for coloured colonies.
CFU/ml= (No of colonies X dilution factor)/ volume of culture on plate
C) E. Coli in Brilliance E. coli agar3
1. Add 25g of food sample to 225ml of 0.1% (w/v) sterile Peptone Water
(CM0009), and stomach for a minimum of 30 seconds to mix the sample
on day1.
2. Incubate the broth at 30ºC for 24 ± 2 hours.
3. On day 2 Gently agitate the bag then, using a microbiological loop,
remove 10μl and inoculate onto an Brilliance E. coli Agar plate and
incubate at 37ºC for 24 ± 2 hours. Count the numbers of pink and purple
colonies
Positive and negative controls
1. Streak a plate of nutrient agar with a glycerol stock of organisms listed as
positive and negative controls in Table-1 on Day 0.
2. On day 1 add one colony to 100 ml of nutrient broth and incubate the broth at
30ºC for 24 ± 2 hours.
3. On day 2 Gently agitate the culture, using a microbiological loop, remove 10μl
and inoculate onto an Brilliance E. coli Agar plate and incubate at 37ºC for 24 ±
2 hours. Examine the plates for pink and purple colonies
CFU/ml= (No of colonies X dilution factor)/ volume of culture on plate
Broth (CM0669), and stomach for a minimum of 30 seconds to mix the
sample on day1.
2. Incubate the broth at 30ºC for 24 ± 2 hours.
3. On day 2 Gently agitate the bag then, using a microbiological loop,
remove 10μl and inoculate onto an Salmonella chromogenic Agar plate
and incubate at 37ºC for 24 ± 2 hours. Examine the plates for coloured
colonies.
Positive and negative controls
1. Streak a plate of nutrient agar with a glycerol stock of organisms listed as
positive and negative controls in Table-1 on Day 0.
2. On day 1 add one colony to 100 ml of nutrient broth and incubate the broth at
30ºC for 24 ± 2 hours.
3. On day 2 Gently agitate the culture, using a microbiological loop, remove 10μl
and inoculate onto an Salmonella chromogenic Agar plate and incubate at 37ºC
for 24 ± 2 hours. Examine the plates for coloured colonies.
CFU/ml= (No of colonies X dilution factor)/ volume of culture on plate
C) E. Coli in Brilliance E. coli agar3
1. Add 25g of food sample to 225ml of 0.1% (w/v) sterile Peptone Water
(CM0009), and stomach for a minimum of 30 seconds to mix the sample
on day1.
2. Incubate the broth at 30ºC for 24 ± 2 hours.
3. On day 2 Gently agitate the bag then, using a microbiological loop,
remove 10μl and inoculate onto an Brilliance E. coli Agar plate and
incubate at 37ºC for 24 ± 2 hours. Count the numbers of pink and purple
colonies
Positive and negative controls
1. Streak a plate of nutrient agar with a glycerol stock of organisms listed as
positive and negative controls in Table-1 on Day 0.
2. On day 1 add one colony to 100 ml of nutrient broth and incubate the broth at
30ºC for 24 ± 2 hours.
3. On day 2 Gently agitate the culture, using a microbiological loop, remove 10μl
and inoculate onto an Brilliance E. coli Agar plate and incubate at 37ºC for 24 ±
2 hours. Examine the plates for pink and purple colonies
CFU/ml= (No of colonies X dilution factor)/ volume of culture on plate
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
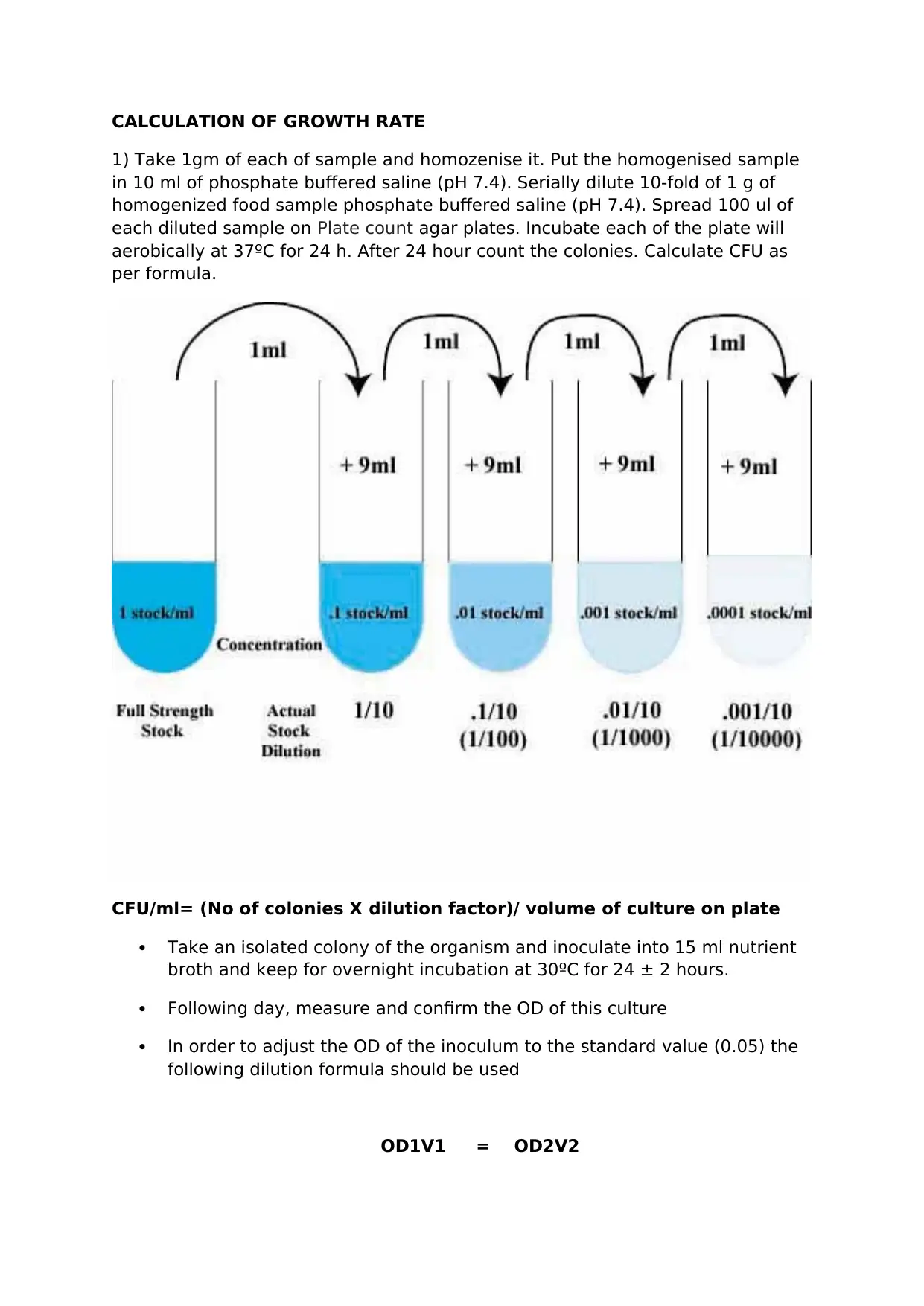
CALCULATION OF GROWTH RATE
1) Take 1gm of each of sample and homozenise it. Put the homogenised sample
in 10 ml of phosphate buffered saline (pH 7.4). Serially dilute 10-fold of 1 g of
homogenized food sample phosphate buffered saline (pH 7.4). Spread 100 ul of
each diluted sample on Plate count agar plates. Incubate each of the plate will
aerobically at 37ºC for 24 h. After 24 hour count the colonies. Calculate CFU as
per formula.
CFU/ml= (No of colonies X dilution factor)/ volume of culture on plate
Take an isolated colony of the organism and inoculate into 15 ml nutrient
broth and keep for overnight incubation at 30ºC for 24 ± 2 hours.
Following day, measure and confirm the OD of this culture
In order to adjust the OD of the inoculum to the standard value (0.05) the
following dilution formula should be used
OD1V1 = OD2V2
1) Take 1gm of each of sample and homozenise it. Put the homogenised sample
in 10 ml of phosphate buffered saline (pH 7.4). Serially dilute 10-fold of 1 g of
homogenized food sample phosphate buffered saline (pH 7.4). Spread 100 ul of
each diluted sample on Plate count agar plates. Incubate each of the plate will
aerobically at 37ºC for 24 h. After 24 hour count the colonies. Calculate CFU as
per formula.
CFU/ml= (No of colonies X dilution factor)/ volume of culture on plate
Take an isolated colony of the organism and inoculate into 15 ml nutrient
broth and keep for overnight incubation at 30ºC for 24 ± 2 hours.
Following day, measure and confirm the OD of this culture
In order to adjust the OD of the inoculum to the standard value (0.05) the
following dilution formula should be used
OD1V1 = OD2V2
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Where,
OD1 = OD of the broth culture, inoculated the previous day.
V1 = volume of this broth culture to be added to the
inoculums
OD2 = OD of the inoculum (as a standard, this value was
adjusted to 0.05)
V2 = volume of the inoculums (in this experiment, 50 ml)
Substitute the values in the equation and calculate V1.
Pipette out that much amount (V1) of the inoculums before adding an
equivalent amount of the broth to it, so that the net volume remains
constant.
Check the O.D at 600nm at every 30 minutes interval and record it.
Plotted using this OD value, a standardized growth curve of the organism
(Absorbance verses time).
Bacterial growth rate will be calculated.
References:
1)http://www.oxoid.com/UK/blue/prod_detail/prod_detail.asp?
pr=CM1080&cat=&c=UK&lang=EN.
2)http://www.oxoid.com/UK/blue/prod_detail/prod_detail.asp?
pr=CM1007&org=124&c=UK&lang=EN.
3) http://www.oxoid.com/uk/blue/prod_detail/prod_detail.asp?pr=cm1046.
OD1 = OD of the broth culture, inoculated the previous day.
V1 = volume of this broth culture to be added to the
inoculums
OD2 = OD of the inoculum (as a standard, this value was
adjusted to 0.05)
V2 = volume of the inoculums (in this experiment, 50 ml)
Substitute the values in the equation and calculate V1.
Pipette out that much amount (V1) of the inoculums before adding an
equivalent amount of the broth to it, so that the net volume remains
constant.
Check the O.D at 600nm at every 30 minutes interval and record it.
Plotted using this OD value, a standardized growth curve of the organism
(Absorbance verses time).
Bacterial growth rate will be calculated.
References:
1)http://www.oxoid.com/UK/blue/prod_detail/prod_detail.asp?
pr=CM1080&cat=&c=UK&lang=EN.
2)http://www.oxoid.com/UK/blue/prod_detail/prod_detail.asp?
pr=CM1007&org=124&c=UK&lang=EN.
3) http://www.oxoid.com/uk/blue/prod_detail/prod_detail.asp?pr=cm1046.
1 out of 5
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.