Programming with R | Assignment
VerifiedAdded on 2022/08/17
|6
|876
|13
AI Summary
Contribute Materials
Your contribution can guide someone’s learning journey. Share your
documents today.

Programming with R
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

Programming with R
The output for the test function in the section in Question 1, produced the result shown in the
table, Table 1: Test function Code and Output below, indicating that the datasets from both
functions used for loading were identical based on the number of rows and columns.
Table 1: Test function Code and Output
test.load.data(Data1,Data2)
[1] "Identical Datasets"
The review process of the data for the section in Question 2, involved creating a function for
checking for null entries and removing them from the dataset to form a new dataset. The second
part of the review of the data involved the check for data points that are too large. This was
achieved by creating a function to check for and omit data points that were upper outliers. The
review process established the presence of outliers but not the presence of values that were too
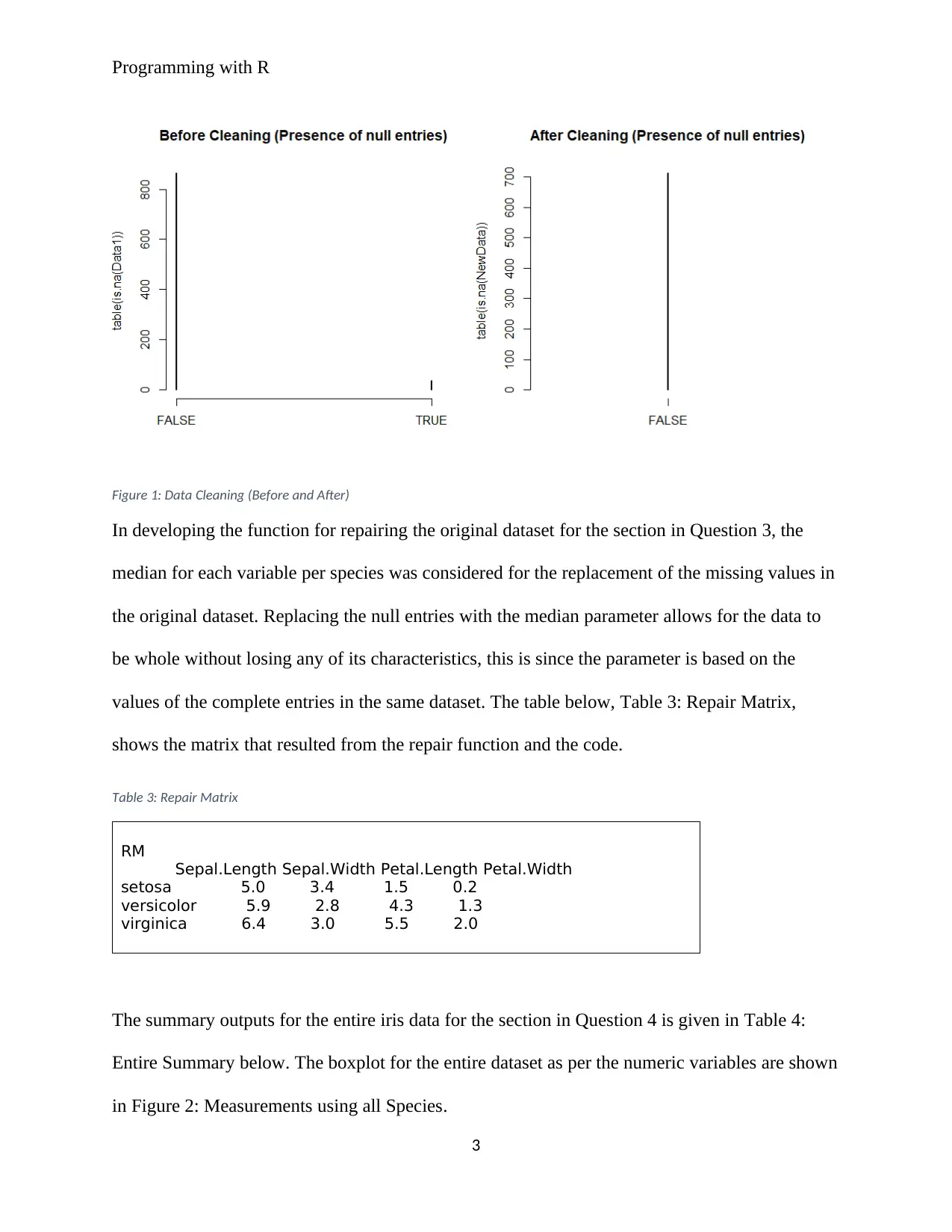
large. The plots below in Figure 1: Data Cleaning (Before and After) shows the before and after
the omission of null entries from the dataset. From the plot, we note that the presence of null
entries was 0 after cleaning. The plots were generated using the codes in Table 2: Data Cleaning
(Before and After) Code below.
Table 2: Data Cleaning (Before and After) Code
#Plotting for Data Cleaning Process
par(mfrow=c(1,2))
plot(table(is.na(Data1)), main = "Before Cleaning (Presence of null entries)")
plot(table(is.na(NewData)), main = "After Cleaning (Presence of null entries)")
2
The output for the test function in the section in Question 1, produced the result shown in the
table, Table 1: Test function Code and Output below, indicating that the datasets from both
functions used for loading were identical based on the number of rows and columns.
Table 1: Test function Code and Output
test.load.data(Data1,Data2)
[1] "Identical Datasets"
The review process of the data for the section in Question 2, involved creating a function for
checking for null entries and removing them from the dataset to form a new dataset. The second
part of the review of the data involved the check for data points that are too large. This was
achieved by creating a function to check for and omit data points that were upper outliers. The
review process established the presence of outliers but not the presence of values that were too
large. The plots below in Figure 1: Data Cleaning (Before and After) shows the before and after
the omission of null entries from the dataset. From the plot, we note that the presence of null
entries was 0 after cleaning. The plots were generated using the codes in Table 2: Data Cleaning
(Before and After) Code below.
Table 2: Data Cleaning (Before and After) Code
#Plotting for Data Cleaning Process
par(mfrow=c(1,2))
plot(table(is.na(Data1)), main = "Before Cleaning (Presence of null entries)")
plot(table(is.na(NewData)), main = "After Cleaning (Presence of null entries)")
2
Programming with R
Figure 1: Data Cleaning (Before and After)
In developing the function for repairing the original dataset for the section in Question 3, the
median for each variable per species was considered for the replacement of the missing values in
the original dataset. Replacing the null entries with the median parameter allows for the data to
be whole without losing any of its characteristics, this is since the parameter is based on the
values of the complete entries in the same dataset. The table below, Table 3: Repair Matrix,
shows the matrix that resulted from the repair function and the code.
Table 3: Repair Matrix
RM
Sepal.Length Sepal.Width Petal.Length Petal.Width
setosa 5.0 3.4 1.5 0.2
versicolor 5.9 2.8 4.3 1.3
virginica 6.4 3.0 5.5 2.0
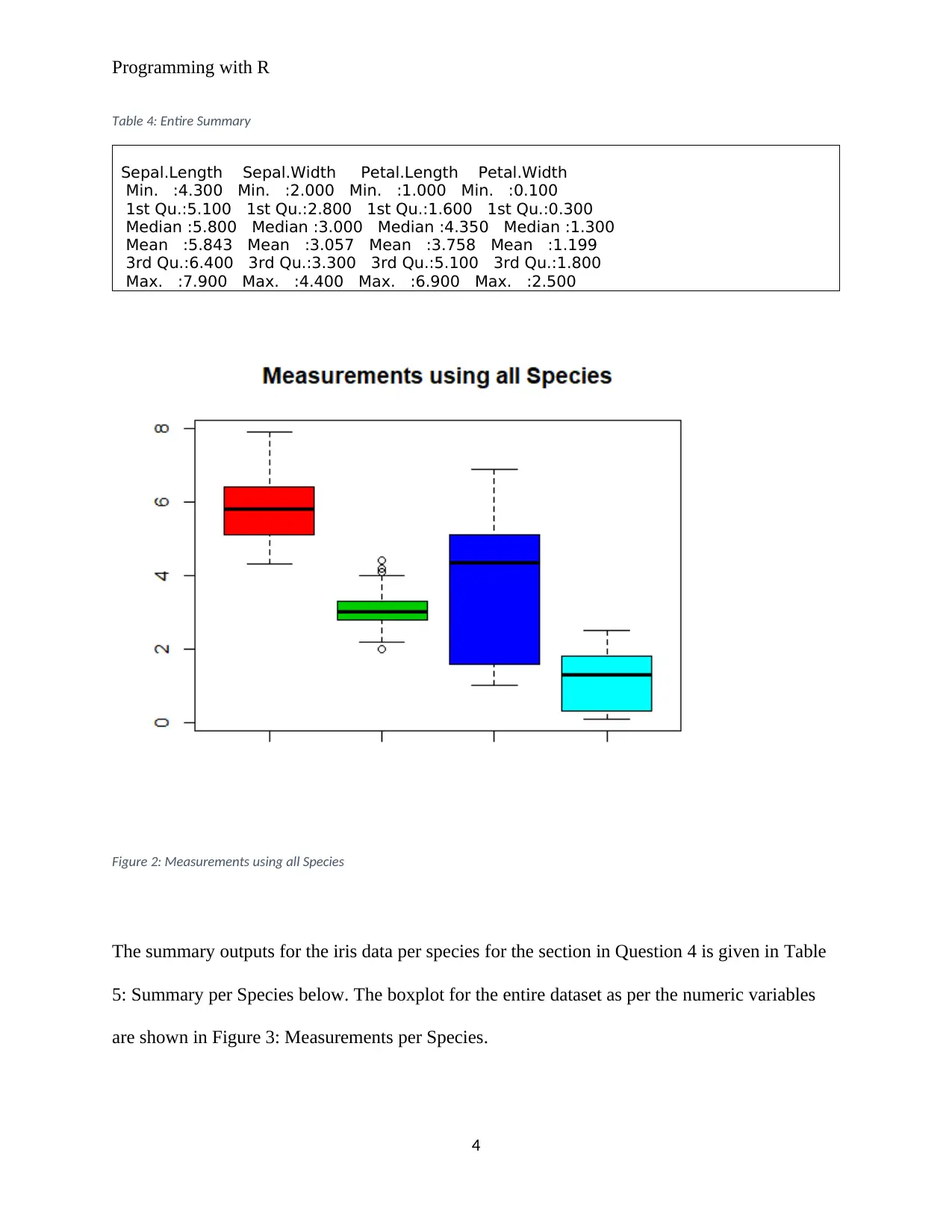
The summary outputs for the entire iris data for the section in Question 4 is given in Table 4:
Entire Summary below. The boxplot for the entire dataset as per the numeric variables are shown
in Figure 2: Measurements using all Species.
3
Figure 1: Data Cleaning (Before and After)
In developing the function for repairing the original dataset for the section in Question 3, the
median for each variable per species was considered for the replacement of the missing values in
the original dataset. Replacing the null entries with the median parameter allows for the data to
be whole without losing any of its characteristics, this is since the parameter is based on the
values of the complete entries in the same dataset. The table below, Table 3: Repair Matrix,
shows the matrix that resulted from the repair function and the code.
Table 3: Repair Matrix
RM
Sepal.Length Sepal.Width Petal.Length Petal.Width
setosa 5.0 3.4 1.5 0.2
versicolor 5.9 2.8 4.3 1.3
virginica 6.4 3.0 5.5 2.0
The summary outputs for the entire iris data for the section in Question 4 is given in Table 4:
Entire Summary below. The boxplot for the entire dataset as per the numeric variables are shown
in Figure 2: Measurements using all Species.
3
Programming with R
Table 4: Entire Summary
Sepal.Length Sepal.Width Petal.Length Petal.Width
Min. :4.300 Min. :2.000 Min. :1.000 Min. :0.100
1st Qu.:5.100 1st Qu.:2.800 1st Qu.:1.600 1st Qu.:0.300
Median :5.800 Median :3.000 Median :4.350 Median :1.300
Mean :5.843 Mean :3.057 Mean :3.758 Mean :1.199
3rd Qu.:6.400 3rd Qu.:3.300 3rd Qu.:5.100 3rd Qu.:1.800
Max. :7.900 Max. :4.400 Max. :6.900 Max. :2.500
Figure 2: Measurements using all Species
The summary outputs for the iris data per species for the section in Question 4 is given in Table
5: Summary per Species below. The boxplot for the entire dataset as per the numeric variables
are shown in Figure 3: Measurements per Species.
4
Table 4: Entire Summary
Sepal.Length Sepal.Width Petal.Length Petal.Width
Min. :4.300 Min. :2.000 Min. :1.000 Min. :0.100
1st Qu.:5.100 1st Qu.:2.800 1st Qu.:1.600 1st Qu.:0.300
Median :5.800 Median :3.000 Median :4.350 Median :1.300
Mean :5.843 Mean :3.057 Mean :3.758 Mean :1.199
3rd Qu.:6.400 3rd Qu.:3.300 3rd Qu.:5.100 3rd Qu.:1.800
Max. :7.900 Max. :4.400 Max. :6.900 Max. :2.500
Figure 2: Measurements using all Species
The summary outputs for the iris data per species for the section in Question 4 is given in Table
5: Summary per Species below. The boxplot for the entire dataset as per the numeric variables
are shown in Figure 3: Measurements per Species.
4
Secure Best Marks with AI Grader
Need help grading? Try our AI Grader for instant feedback on your assignments.

Programming with R
Table 5: Summary per Species
> summaryFunction(iris)
$setosa
Sepal.Length Sepal.Width Petal.Length Petal.Width
Min. :4.300 Min. :2.300 Min. :1.000 Min. :0.100
1st Qu.:4.800 1st Qu.:3.200 1st Qu.:1.400 1st Qu.:0.200
Median :5.000 Median :3.400 Median :1.500 Median :0.200
Mean :5.006 Mean :3.428 Mean :1.462 Mean :0.246
3rd Qu.:5.200 3rd Qu.:3.675 3rd Qu.:1.575 3rd Qu.:0.300
Max. :5.800 Max. :4.400 Max. :1.900 Max. :0.600
$versicolor
Sepal.Length Sepal.Width Petal.Length Petal.Width
Min. :4.900 Min. :2.000 Min. :3.00 Min. :1.000
1st Qu.:5.600 1st Qu.:2.525 1st Qu.:4.00 1st Qu.:1.200
Median :5.900 Median :2.800 Median :4.35 Median :1.300
Mean :5.936 Mean :2.770 Mean :4.26 Mean :1.326
3rd Qu.:6.300 3rd Qu.:3.000 3rd Qu.:4.60 3rd Qu.:1.500
Max. :7.000 Max. :3.400 Max. :5.10 Max. :1.800
$virginica
Sepal.Length Sepal.Width Petal.Length Petal.Width
Min. :4.900 Min. :2.200 Min. :4.500 Min. :1.400
1st Qu.:6.225 1st Qu.:2.800 1st Qu.:5.100 1st Qu.:1.800
Median :6.500 Median :3.000 Median :5.550 Median :2.000
Mean :6.588 Mean :2.974 Mean :5.552 Mean :2.026
3rd Qu.:6.900 3rd Qu.:3.175 3rd Qu.:5.875 3rd Qu.:2.300
Max. :7.900 Max. :3.800 Max. :6.900 Max. :2.500
5
Table 5: Summary per Species
> summaryFunction(iris)
$setosa
Sepal.Length Sepal.Width Petal.Length Petal.Width
Min. :4.300 Min. :2.300 Min. :1.000 Min. :0.100
1st Qu.:4.800 1st Qu.:3.200 1st Qu.:1.400 1st Qu.:0.200
Median :5.000 Median :3.400 Median :1.500 Median :0.200
Mean :5.006 Mean :3.428 Mean :1.462 Mean :0.246
3rd Qu.:5.200 3rd Qu.:3.675 3rd Qu.:1.575 3rd Qu.:0.300
Max. :5.800 Max. :4.400 Max. :1.900 Max. :0.600
$versicolor
Sepal.Length Sepal.Width Petal.Length Petal.Width
Min. :4.900 Min. :2.000 Min. :3.00 Min. :1.000
1st Qu.:5.600 1st Qu.:2.525 1st Qu.:4.00 1st Qu.:1.200
Median :5.900 Median :2.800 Median :4.35 Median :1.300
Mean :5.936 Mean :2.770 Mean :4.26 Mean :1.326
3rd Qu.:6.300 3rd Qu.:3.000 3rd Qu.:4.60 3rd Qu.:1.500
Max. :7.000 Max. :3.400 Max. :5.10 Max. :1.800
$virginica
Sepal.Length Sepal.Width Petal.Length Petal.Width
Min. :4.900 Min. :2.200 Min. :4.500 Min. :1.400
1st Qu.:6.225 1st Qu.:2.800 1st Qu.:5.100 1st Qu.:1.800
Median :6.500 Median :3.000 Median :5.550 Median :2.000
Mean :6.588 Mean :2.974 Mean :5.552 Mean :2.026
3rd Qu.:6.900 3rd Qu.:3.175 3rd Qu.:5.875 3rd Qu.:2.300
Max. :7.900 Max. :3.800 Max. :6.900 Max. :2.500
5

Programming with R
Figure 3: Measurements per Species
6
Figure 3: Measurements per Species
6
1 out of 6
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
© 2024 | Zucol Services PVT LTD | All rights reserved.
