Microbiology Report: Identification of S. epidermidis and Treatment
VerifiedAdded on 2023/05/28
|8
|1912
|51
Report
AI Summary
This microbiology report details the identification and treatment of a Staphylococcus epidermidis infection in a 56-year-old patient. The study employed phenotypic (Gram staining, colony morphology, catalase, coagulase tests) and biochemical methods along with molecular techniques (PCR, electrophoresis, and sequencing) to identify the bacteria. Antibiotic susceptibility testing was performed to determine effective treatments, revealing resistance to certain antibiotics. The report discusses the methods used, the results obtained, and the recommended treatments, including Gentamycin, Linezolid, and Vancomycin, considering the patient's clinical condition and antibiotic resistance profiles. The findings highlight the importance of accurate bacterial identification and antibiotic susceptibility testing in guiding effective clinical treatment strategies for infections like those caused by S. epidermidis. The patient was admitted to the ICU following a road traffic accident and developed pyrexia, rigors, and hypotension after catheterization, prompting the investigation.

MICROBIOLOGY
By Name
Course
Instructor
Institution
Location
Date
Treatment of the clinical Signs and Symptoms through Identification and Isolation mechanism of suspected
bacteria
By Name
Course
Instructor
Institution
Location
Date
Treatment of the clinical Signs and Symptoms through Identification and Isolation mechanism of suspected
bacteria
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

INTRODUCTION
A 56-year-old man who was actually catherized became pyrexic, showed rigors and hypotension just eight days
after his admission to an ICU with the injuries that resulted from the road accident. In order to facilitate the
process of the treatment, the blood samples were taken from his venous line then introduce into the incubation
bottles that were sent to the laboratory of the micro-biology where they were incubated using automated
incubation machine(Paul 2014). The samples were flanged positive the following day but the subsequent gram-
stain was actually found to be negative on testing. Coagulase test further narrowed the identification down as it
produced a negative result, meaning that patient was not infected with Staphylococcus aureus, but rather with
coagulase-negative bacteria. Staphylococcus Epidermidis.
Methods
Identification through phenotypic and biochemical means
There was use of the gram staining mechanism for both positive and negative results. The slide was flooded with
the gram stain 1 and then left for 1 minute. The stains were then washed off with the water and slide flooded with
the gram stain2 for another 1minute.It was washed in water then blot dry.Decolorization was carried out for
30minutes by flooding with95%alcohol.
It was executed as portrayed by scholars (Paul 2014) Next, microscopic organisms were refined on 2 unique kinds
of specific media - Baird Parker agar, which is specific for Staphylococci which separates microorganisms,
especially Staphylococci, in light of the deoxyribonuclease creation(Markey, Leonard, Archambault, Cullinane
and Maguire 2013). A short time later, catalase test was performed to research whether bubble arrangement
happens upon contact among microscopic organisms and hydrogen peroxide because of catalase chemical
creation(Paul 2014) As to coagulase testing, it was performed by latex agglutination on a card so as to set up
whether the microorganisms are creating a coagulase protein(Wood 2012).Living being distinguishing proof at an
A 56-year-old man who was actually catherized became pyrexic, showed rigors and hypotension just eight days
after his admission to an ICU with the injuries that resulted from the road accident. In order to facilitate the
process of the treatment, the blood samples were taken from his venous line then introduce into the incubation
bottles that were sent to the laboratory of the micro-biology where they were incubated using automated
incubation machine(Paul 2014). The samples were flanged positive the following day but the subsequent gram-
stain was actually found to be negative on testing. Coagulase test further narrowed the identification down as it
produced a negative result, meaning that patient was not infected with Staphylococcus aureus, but rather with
coagulase-negative bacteria. Staphylococcus Epidermidis.
Methods
Identification through phenotypic and biochemical means
There was use of the gram staining mechanism for both positive and negative results. The slide was flooded with
the gram stain 1 and then left for 1 minute. The stains were then washed off with the water and slide flooded with
the gram stain2 for another 1minute.It was washed in water then blot dry.Decolorization was carried out for
30minutes by flooding with95%alcohol.
It was executed as portrayed by scholars (Paul 2014) Next, microscopic organisms were refined on 2 unique kinds
of specific media - Baird Parker agar, which is specific for Staphylococci which separates microorganisms,
especially Staphylococci, in light of the deoxyribonuclease creation(Markey, Leonard, Archambault, Cullinane
and Maguire 2013). A short time later, catalase test was performed to research whether bubble arrangement
happens upon contact among microscopic organisms and hydrogen peroxide because of catalase chemical
creation(Paul 2014) As to coagulase testing, it was performed by latex agglutination on a card so as to set up
whether the microorganisms are creating a coagulase protein(Wood 2012).Living being distinguishing proof at an

organism categories level was accomplished with the assistance of Microbact 12S Staphylococcal Identification
System.
Identification by molecular means
DNA was separated by boiling and centrifuging the example and after that Polymerase Chain Reaction (PCR)
blend was set up so as to enhance DNA of enthusiasm for nucleotide succession correlation. The 0.8 kb PCR-
intensified 16S rRNA quality parts were utilized as a sequencing layout close by with 8FPL and 806R
preliminaries. PCR results were confirmed by electrophoresis, which isolates DNA pieces dependent on their size
(Wood 2012). Phosphate bunches in DNA spine are contrarily charged, along these lines DNA pieces are pulled in
to the positive charge and the littler they are, the quicker they move through agarose gel(Paul 2014).The DNA
sections in the example were tried against the DNA parts of known length (stepping stool). Afterward, the
acquired outcomes were utilized to distinguish the grouping root by utilizing Basic Local Alignment Search Tool.
Suggested Methods for treatments
So as to build up potential treatment for the patient, antimicrobial testing was performed. Anti-toxin rings were
put on the plates with microorganisms to screen the bacterial development hindrance by different anti-infection
agents. In addition, Vancomycin Minimum Inhibitory Concentration (MIC) was resolved so as to test for bacterial
weakness to Vancomycin and to set up the base focus at which microbes are repressed.
RESULTS AND DISCUSSION
Biochemical and Phenotypic identification means
While surveying state morphology, it was discovered that white-colored level bunched settlements have
developed on the blood agar. Refined on particular media brought about bacterial development on Baird-Parker
yet not on DNase agar, meaning the absence of DNase action (Paul 2014) The outcomes for particular
System.
Identification by molecular means
DNA was separated by boiling and centrifuging the example and after that Polymerase Chain Reaction (PCR)
blend was set up so as to enhance DNA of enthusiasm for nucleotide succession correlation. The 0.8 kb PCR-
intensified 16S rRNA quality parts were utilized as a sequencing layout close by with 8FPL and 806R
preliminaries. PCR results were confirmed by electrophoresis, which isolates DNA pieces dependent on their size
(Wood 2012). Phosphate bunches in DNA spine are contrarily charged, along these lines DNA pieces are pulled in
to the positive charge and the littler they are, the quicker they move through agarose gel(Paul 2014).The DNA
sections in the example were tried against the DNA parts of known length (stepping stool). Afterward, the
acquired outcomes were utilized to distinguish the grouping root by utilizing Basic Local Alignment Search Tool.
Suggested Methods for treatments
So as to build up potential treatment for the patient, antimicrobial testing was performed. Anti-toxin rings were
put on the plates with microorganisms to screen the bacterial development hindrance by different anti-infection
agents. In addition, Vancomycin Minimum Inhibitory Concentration (MIC) was resolved so as to test for bacterial
weakness to Vancomycin and to set up the base focus at which microbes are repressed.
RESULTS AND DISCUSSION
Biochemical and Phenotypic identification means
While surveying state morphology, it was discovered that white-colored level bunched settlements have
developed on the blood agar. Refined on particular media brought about bacterial development on Baird-Parker
yet not on DNase agar, meaning the absence of DNase action (Paul 2014) The outcomes for particular
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

development were suggestive of microscopic organisms being Staphylococci and this was additionally affirmed
by the Gram staining. Talaro and Chess 2018 explain that Gram-negative microscopic organisms have a thin
peptidoglycan layer basically made of lipopolysaccharides and lipoproteins, while Gram-positive microorganisms
have a thick peptidoglycan layer which fills in as the external film (Ray and Bhunia 2013).
For this situation think about, Gram staining created a positive outcome as bacterial cell walls were seen to be
stained purple under the light magnifying lens. According to Ray and Bhunia 2013,it is with respect to catalase
test which is utilized to separate among Staphylococci and Streptococci, it has given a positive outcome implying
that the detached microscopic organisms were Staphylococci(Banwart 2012) Coagulase test has additionally
limited the recognizable proof down as it delivered a negative outcome, implying that tolerant was not tainted
with Staphylococcus aureus, but instead with coagulase-negative microscopic organisms. Wood 2012 confirms
Microbact 12S Staphylococcal Identification System was utilized to distinguish the living being at an animal
categories level dependent on sugar use and colorimetric catalyst location(Zabel and Morrell 2012) . An inquiry
was kept running in a database for the code acquired from that test for species ID and has given a 99.8%
likelihood that the species took a gander at was Staphylococcus epidermidis (S. epidermidis), which was most
presumably doctor's facility obtained as that is normally the situation with S. epidermidis contaminations and was
probably a skin contaminant (Tille 2015).
Identification via molecular means
Electrophoresis has uncovered that DNA groups for S. epidermidis positive control and the tried example were at
a similar locus, implying that the DNA parts were of a similar length. The nucleotide succession got from
sequencing was broke down utilizing BLAST so as to decide the species. The pursuit brought about 165 BLAST
hits on the question succession, 30 of which were for S.epidermidis and the other 135 for Staphylococcus species
or for uncultured bacterium incomplete 16S rRNA quality. E esteem was 0 for each BLAST hit, implying that the
matches were immaculate and not because of the possibility arrangement. In addition, it implied that the hit
sequence coordinated the inquiry arrangement over its full length (Doyle and Buchanan 2012).
by the Gram staining. Talaro and Chess 2018 explain that Gram-negative microscopic organisms have a thin
peptidoglycan layer basically made of lipopolysaccharides and lipoproteins, while Gram-positive microorganisms
have a thick peptidoglycan layer which fills in as the external film (Ray and Bhunia 2013).
For this situation think about, Gram staining created a positive outcome as bacterial cell walls were seen to be
stained purple under the light magnifying lens. According to Ray and Bhunia 2013,it is with respect to catalase
test which is utilized to separate among Staphylococci and Streptococci, it has given a positive outcome implying
that the detached microscopic organisms were Staphylococci(Banwart 2012) Coagulase test has additionally
limited the recognizable proof down as it delivered a negative outcome, implying that tolerant was not tainted
with Staphylococcus aureus, but instead with coagulase-negative microscopic organisms. Wood 2012 confirms
Microbact 12S Staphylococcal Identification System was utilized to distinguish the living being at an animal
categories level dependent on sugar use and colorimetric catalyst location(Zabel and Morrell 2012) . An inquiry
was kept running in a database for the code acquired from that test for species ID and has given a 99.8%
likelihood that the species took a gander at was Staphylococcus epidermidis (S. epidermidis), which was most
presumably doctor's facility obtained as that is normally the situation with S. epidermidis contaminations and was
probably a skin contaminant (Tille 2015).
Identification via molecular means
Electrophoresis has uncovered that DNA groups for S. epidermidis positive control and the tried example were at
a similar locus, implying that the DNA parts were of a similar length. The nucleotide succession got from
sequencing was broke down utilizing BLAST so as to decide the species. The pursuit brought about 165 BLAST
hits on the question succession, 30 of which were for S.epidermidis and the other 135 for Staphylococcus species
or for uncultured bacterium incomplete 16S rRNA quality. E esteem was 0 for each BLAST hit, implying that the
matches were immaculate and not because of the possibility arrangement. In addition, it implied that the hit
sequence coordinated the inquiry arrangement over its full length (Doyle and Buchanan 2012).
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
Suggested treatment methods
Results for treatment recommendations
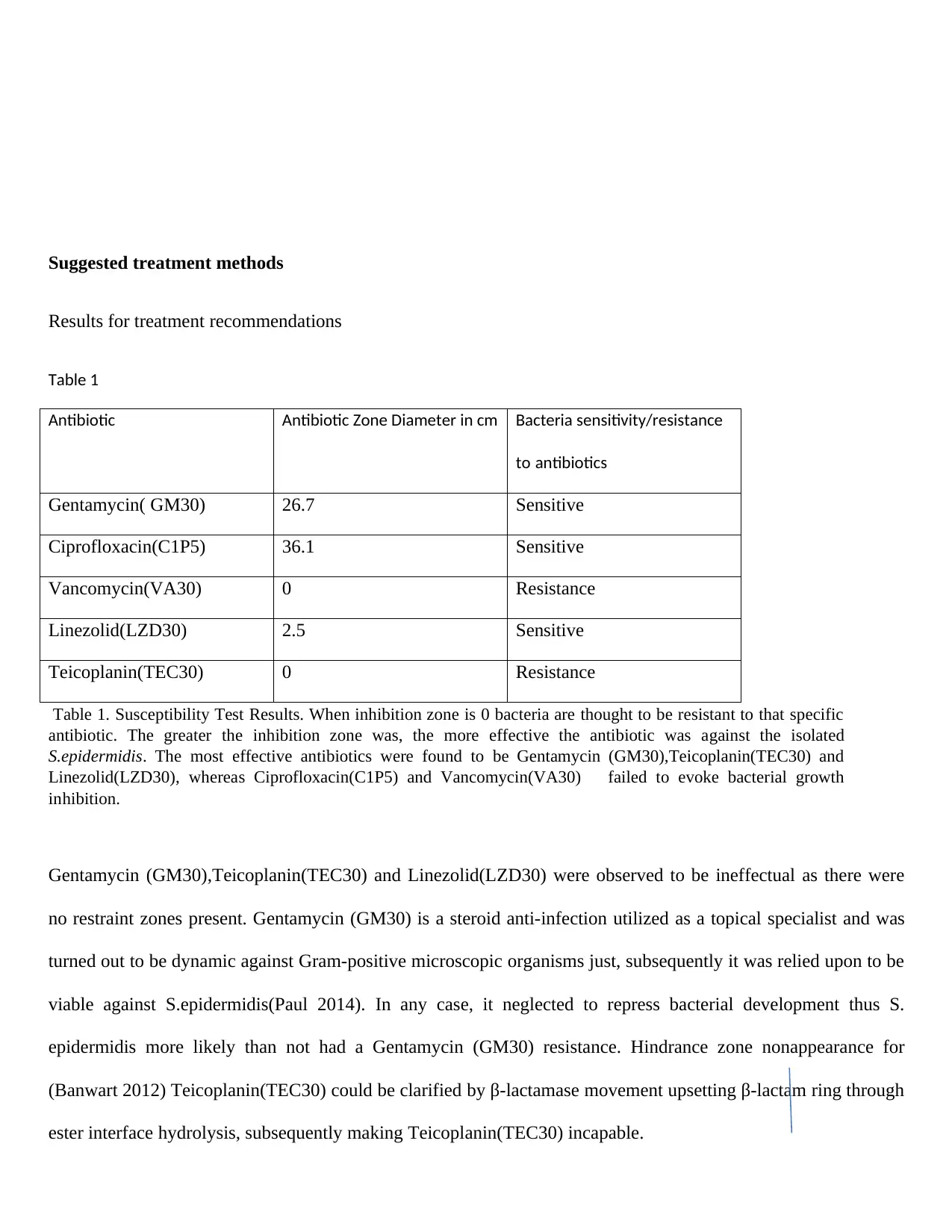
Table 1
Antibiotic Antibiotic Zone Diameter in cm Bacteria sensitivity/resistance
to antibiotics
Gentamycin( GM30) 26.7 Sensitive
Ciprofloxacin(C1P5) 36.1 Sensitive
Vancomycin(VA30) 0 Resistance
Linezolid(LZD30) 2.5 Sensitive
Teicoplanin(TEC30) 0 Resistance
Table 1. Susceptibility Test Results. When inhibition zone is 0 bacteria are thought to be resistant to that specific
antibiotic. The greater the inhibition zone was, the more effective the antibiotic was against the isolated
S.epidermidis. The most effective antibiotics were found to be Gentamycin (GM30),Teicoplanin(TEC30) and
Linezolid(LZD30), whereas Ciprofloxacin(C1P5) and Vancomycin(VA30) failed to evoke bacterial growth
inhibition.
Gentamycin (GM30),Teicoplanin(TEC30) and Linezolid(LZD30) were observed to be ineffectual as there were
no restraint zones present. Gentamycin (GM30) is a steroid anti-infection utilized as a topical specialist and was
turned out to be dynamic against Gram-positive microscopic organisms just, subsequently it was relied upon to be
viable against S.epidermidis(Paul 2014). In any case, it neglected to repress bacterial development thus S.
epidermidis more likely than not had a Gentamycin (GM30) resistance. Hindrance zone nonappearance for
(Banwart 2012) Teicoplanin(TEC30) could be clarified by β-lactamase movement upsetting β-lactam ring through
ester interface hydrolysis, subsequently making Teicoplanin(TEC30) incapable.
Results for treatment recommendations
Table 1
Antibiotic Antibiotic Zone Diameter in cm Bacteria sensitivity/resistance
to antibiotics
Gentamycin( GM30) 26.7 Sensitive
Ciprofloxacin(C1P5) 36.1 Sensitive
Vancomycin(VA30) 0 Resistance
Linezolid(LZD30) 2.5 Sensitive
Teicoplanin(TEC30) 0 Resistance
Table 1. Susceptibility Test Results. When inhibition zone is 0 bacteria are thought to be resistant to that specific
antibiotic. The greater the inhibition zone was, the more effective the antibiotic was against the isolated
S.epidermidis. The most effective antibiotics were found to be Gentamycin (GM30),Teicoplanin(TEC30) and
Linezolid(LZD30), whereas Ciprofloxacin(C1P5) and Vancomycin(VA30) failed to evoke bacterial growth
inhibition.
Gentamycin (GM30),Teicoplanin(TEC30) and Linezolid(LZD30) were observed to be ineffectual as there were
no restraint zones present. Gentamycin (GM30) is a steroid anti-infection utilized as a topical specialist and was
turned out to be dynamic against Gram-positive microscopic organisms just, subsequently it was relied upon to be
viable against S.epidermidis(Paul 2014). In any case, it neglected to repress bacterial development thus S.
epidermidis more likely than not had a Gentamycin (GM30) resistance. Hindrance zone nonappearance for
(Banwart 2012) Teicoplanin(TEC30) could be clarified by β-lactamase movement upsetting β-lactam ring through
ester interface hydrolysis, subsequently making Teicoplanin(TEC30) incapable.
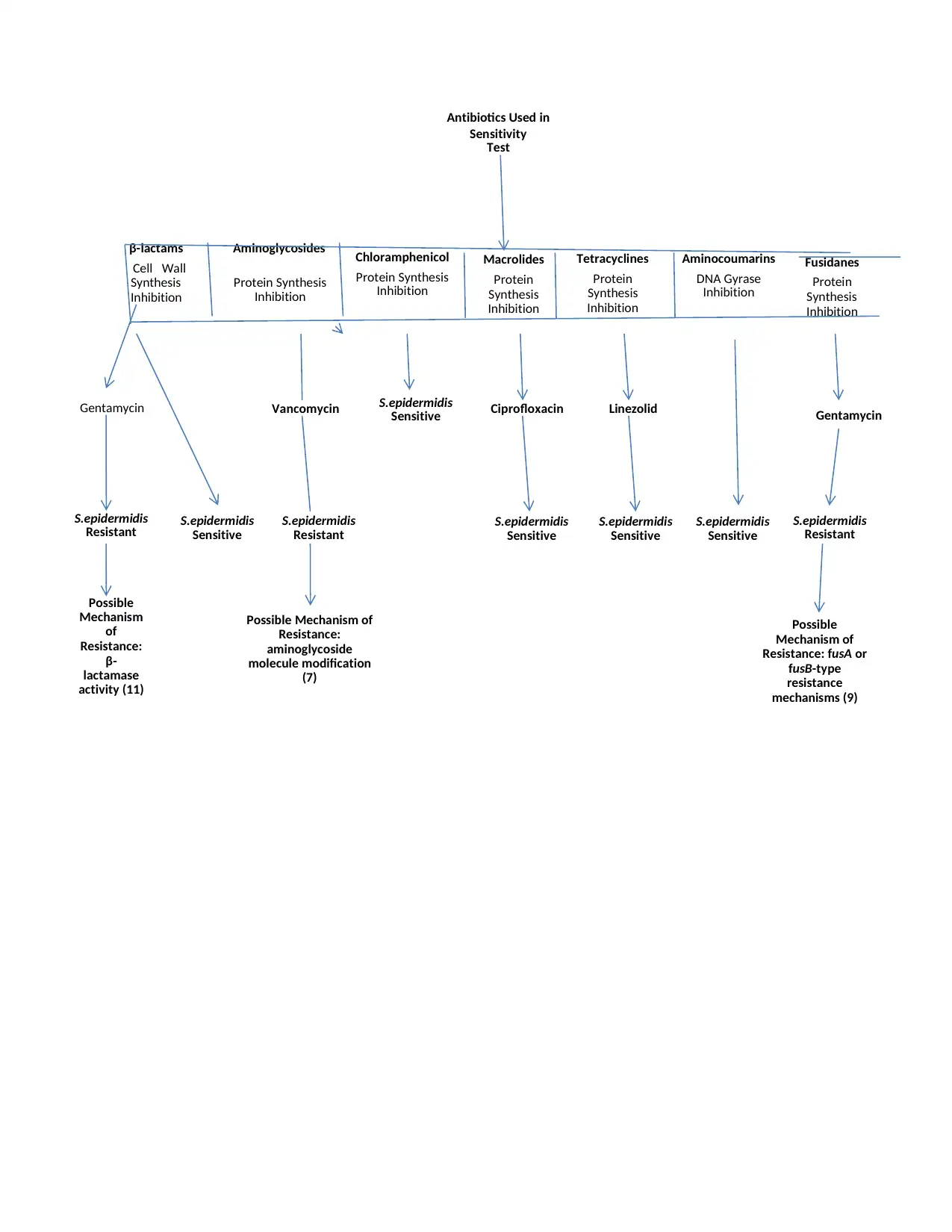
Antibiotics Used in
Sensitivity
Test
β-lactams
Cell Wall
Synthesis
Inhibition
Aminoglycosides
Protein Synthesis
Inhibition
Chloramphenicol
Protein Synthesis
Inhibition
Macrolides
Protein
Synthesis
Inhibition
Tetracyclines
Protein
Synthesis
Inhibition
Aminocoumarins
DNA Gyrase
Inhibition
Fusidanes
Protein
Synthesis
Inhibition
Gentamycin Vancomycin S.epidermidis
Sensitive Ciprofloxacin Linezolid Gentamycin
S.epidermidis
Resistant S.epidermidis
Sensitive
S.epidermidis
Resistant S.epidermidis
Sensitive
S.epidermidis
Sensitive
S.epidermidis
Sensitive
S.epidermidis
Resistant
Possible
Mechanism
of
Resistance:
β-
lactamase
activity (11)
Possible Mechanism of
Resistance:
aminoglycoside
molecule modification
(7)
Possible
Mechanism of
Resistance: fusA or
fusB-type
resistance
mechanisms (9)
Sensitivity
Test
β-lactams
Cell Wall
Synthesis
Inhibition
Aminoglycosides
Protein Synthesis
Inhibition
Chloramphenicol
Protein Synthesis
Inhibition
Macrolides
Protein
Synthesis
Inhibition
Tetracyclines
Protein
Synthesis
Inhibition
Aminocoumarins
DNA Gyrase
Inhibition
Fusidanes
Protein
Synthesis
Inhibition
Gentamycin Vancomycin S.epidermidis
Sensitive Ciprofloxacin Linezolid Gentamycin
S.epidermidis
Resistant S.epidermidis
Sensitive
S.epidermidis
Resistant S.epidermidis
Sensitive
S.epidermidis
Sensitive
S.epidermidis
Sensitive
S.epidermidis
Resistant
Possible
Mechanism
of
Resistance:
β-
lactamase
activity (11)
Possible Mechanism of
Resistance:
aminoglycoside
molecule modification
(7)
Possible
Mechanism of
Resistance: fusA or
fusB-type
resistance
mechanisms (9)
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

As to, it is an aminoglycoside which is once in a while utilized in clinical settings because of a
fast obstruction improvement by microscopic organisms. Most likely S. epidermidis confined
from the patient was impervious to Streptomycin (Talaro and Chess 2018). Ciprofloxacin(C1P5)
caused restraint in any case; it was not as viable as, Vancomycin(VA30). Ciprofloxacin(C1P5)
has a place with isoxazolyl gathering of Teicoplanin(TEC30), which has β-lactam security
shielded from β-lactamase action because of a tight substrate particularity shown by β-
lactamases, and it was demonstrated that Staphylococal β-lactamases don't perceive isoxazolyls
as substrates(Paul 2014) This could clear up why disconnected S. epidermidis was impervious to
Teicoplanin(TEC30) however was as yet restrained by Ciprofloxacin(C1P5). Anti-toxins talked
about above did not share anti-microbial family or the instrument of activity, in this way
disengaged S. epidermidis microorganisms were multi-medicate safe(Tille 2015)
Vancomycin Minimum Inhibitory Concentration (MIC) was built up to be 2 μg/ml which
implied this was the base focus at which Vancomycin had the capacity to hinder bacterial
development; accordingly it would be viable in treating the patient.
CONCLUSION
Through the application of various strategies, for example, particular refined, biochemical tests
and PCR joined with electrophoresis, it was built up that tolerant was tainted with S. epidermidis.
By exploring the affectability of this species towards different anti-toxins it was discovered that
patient could be treated with either Gentamycin (GM30), Teicoplanin(TEC30) and
Linezolid(LZD30) or Vancomycin(VA30). Because of danger and simplicity of opposition
fast obstruction improvement by microscopic organisms. Most likely S. epidermidis confined
from the patient was impervious to Streptomycin (Talaro and Chess 2018). Ciprofloxacin(C1P5)
caused restraint in any case; it was not as viable as, Vancomycin(VA30). Ciprofloxacin(C1P5)
has a place with isoxazolyl gathering of Teicoplanin(TEC30), which has β-lactam security
shielded from β-lactamase action because of a tight substrate particularity shown by β-
lactamases, and it was demonstrated that Staphylococal β-lactamases don't perceive isoxazolyls
as substrates(Paul 2014) This could clear up why disconnected S. epidermidis was impervious to
Teicoplanin(TEC30) however was as yet restrained by Ciprofloxacin(C1P5). Anti-toxins talked
about above did not share anti-microbial family or the instrument of activity, in this way
disengaged S. epidermidis microorganisms were multi-medicate safe(Tille 2015)
Vancomycin Minimum Inhibitory Concentration (MIC) was built up to be 2 μg/ml which
implied this was the base focus at which Vancomycin had the capacity to hinder bacterial
development; accordingly it would be viable in treating the patient.
CONCLUSION
Through the application of various strategies, for example, particular refined, biochemical tests
and PCR joined with electrophoresis, it was built up that tolerant was tainted with S. epidermidis.
By exploring the affectability of this species towards different anti-toxins it was discovered that
patient could be treated with either Gentamycin (GM30), Teicoplanin(TEC30) and
Linezolid(LZD30) or Vancomycin(VA30). Because of danger and simplicity of opposition
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

advancement of the greater part of them, Erythromycin or Vancomycin would be an ideal
treatment for this situation.
BIBLIOGRAPHY
Banwart, G., 2012. Basic food microbiology. Springer Science & Business Media.
Doyle, M.P. and Buchanan, R.L. eds., 2012. Food microbiology: fundamentals and frontiers.
American Society for Microbiology Press.
Markey, B., Leonard, F., Archambault, M., Cullinane, A. and Maguire, D., 2013. Clinical
Veterinary Microbiology E-Book. Elsevier Health Sciences.
Paul, E.A., 2014. Soil microbiology, ecology and biochemistry. Academic press.
Ray, B. and Bhunia, A., 2013. Fundamental food microbiology. CRC press.
Talaro, K.P. and Chess, B., 2018. Foundations in microbiology. McGraw-Hill.
Tille, P., 2015. Bailey & Scott's Diagnostic Microbiology-E-Book. Elsevier Health Sciences.
Wood, B.J., 2012. Microbiology of fermented foods. Springer Science & Business Media.
Zabel, R.A. and Morrell, J.J., 2012. Wood microbiology: decay and its prevention. Academic
press.
treatment for this situation.
BIBLIOGRAPHY
Banwart, G., 2012. Basic food microbiology. Springer Science & Business Media.
Doyle, M.P. and Buchanan, R.L. eds., 2012. Food microbiology: fundamentals and frontiers.
American Society for Microbiology Press.
Markey, B., Leonard, F., Archambault, M., Cullinane, A. and Maguire, D., 2013. Clinical
Veterinary Microbiology E-Book. Elsevier Health Sciences.
Paul, E.A., 2014. Soil microbiology, ecology and biochemistry. Academic press.
Ray, B. and Bhunia, A., 2013. Fundamental food microbiology. CRC press.
Talaro, K.P. and Chess, B., 2018. Foundations in microbiology. McGraw-Hill.
Tille, P., 2015. Bailey & Scott's Diagnostic Microbiology-E-Book. Elsevier Health Sciences.
Wood, B.J., 2012. Microbiology of fermented foods. Springer Science & Business Media.
Zabel, R.A. and Morrell, J.J., 2012. Wood microbiology: decay and its prevention. Academic
press.
1 out of 8
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.

