Bioinformatics Analysis: Mycoplasma genitalium Protein Sequence
VerifiedAdded on 2023/04/21
|22
|4004
|416
Report
AI Summary
This report presents a bioinformatics analysis of a Mycoplasma genitalium protein sequence, utilizing tools like BLAST-p, PSI-BLAST, FASTA, PROSITE, Pfam, EBI, and SWISS PROT to assess its structural and functional characteristics. The analysis identified the sequence as belonging to the MG045 solute binding family, highlighting its role in polyamine uptake and periplasmic binding. The study revealed the protein's homology with other known sequences and its capacity for structural modification in response to cellular needs. Results from the Ramachandran Plot indicated structural consistency. The report explores the protein's conserved domains, including Lipoprotein_8, PotD, and PBP2_PotD_PotF_like components, and discusses the implications of these findings in relation to the organism's pathogenicity and metabolic processes. The analysis also investigates the sequence's relationship with other organisms. The report concludes by acknowledging the limitations of the study while emphasizing its contribution to understanding the biological structure and function of the analyzed protein sequence.

P a g e | 1
Abstract
The presented paper evaluated the sequence related to Mycoplasma genitalium through the
utilization of various bioinformatics tools, methods, and databases. BLAST-p, PSI-BLAST,
FASTA, PROSITE, Pfam, EBI, and SWISS PROT include some of the significant tools that
were utilized in the paper for retrieving the protein variant of M. genitalium and comparing
the same with other protein structures. The structural and functional assessment of the protein
structure assisted in predicting the consistency and engagement of M. genitalium protein in
various biological activities. The provided sequence of M. genitalium pertained to the
MG045 solute binding family. The retrieved protein structure was found to be related to the
significant processes including polyamines uptake and periplasmic binding. The results also
revealed the smallest structure of M. genitalium protein as compared to the structures of other
free-living organisms. The presented structure exhibited substantial homology with other
known protein sequences of the same organism. The multiple alignments of M. genitalium
protein sequence with a randomly selected sequence also revealed substantial similarity in
some of the selected base pairs. The secondary structures of M. genitalium revealed 12-23%
sequence identity. The structural assessment revealed the high capacity of M. genitalium
proteins to modify themselves in accordance with the cellular metabolic requirements. 83%-
85% scores revealed from the Ramachandran Plot demonstrated the structural consistency of
M. genitalium protein. However, a range of M. genitalium variants still requires a structural
and functional assessment to evaluate their biological potential. The present findings from the
existing bioinformatics tool despite their accuracy might not be generalized across the wider
genome pool of M. genitalium because of their limited scalability. However, the study has
substantially enhanced the insight regarding the biological structure and function of the
presented M. genitalium protein sequence.
Introduction
The genital mycoplasmas categorize Mycoplasma genitalium as a microbe that leads to the
development of sexually transmitted infections in the human population (Sethi, Singh,
Samanta, & Sharma, 2012). Evidence-based literature describes the flask-shaped morphology
of Mycoplasma genitalium based on its terminal orgallene (i.e. cell membrane protrusion).
This organelle is a virulence factor that facilitates the motility and adhesion of Mycoplasma
genitalium across the host cell line. The cytoskeleton of the terminal organelle of
Mycoplasma genitalium corresponds to 300nm and incorporates the wheel complex, rod, and
terminal button (Martinelli et al., 2016). The crystal structure of Mycoplasma genitalium
protein (MG491) is based on a resolution of 3-angstrom. The subunits of Mycoplasma
Abstract
The presented paper evaluated the sequence related to Mycoplasma genitalium through the
utilization of various bioinformatics tools, methods, and databases. BLAST-p, PSI-BLAST,
FASTA, PROSITE, Pfam, EBI, and SWISS PROT include some of the significant tools that
were utilized in the paper for retrieving the protein variant of M. genitalium and comparing
the same with other protein structures. The structural and functional assessment of the protein
structure assisted in predicting the consistency and engagement of M. genitalium protein in
various biological activities. The provided sequence of M. genitalium pertained to the
MG045 solute binding family. The retrieved protein structure was found to be related to the
significant processes including polyamines uptake and periplasmic binding. The results also
revealed the smallest structure of M. genitalium protein as compared to the structures of other
free-living organisms. The presented structure exhibited substantial homology with other
known protein sequences of the same organism. The multiple alignments of M. genitalium
protein sequence with a randomly selected sequence also revealed substantial similarity in
some of the selected base pairs. The secondary structures of M. genitalium revealed 12-23%
sequence identity. The structural assessment revealed the high capacity of M. genitalium
proteins to modify themselves in accordance with the cellular metabolic requirements. 83%-
85% scores revealed from the Ramachandran Plot demonstrated the structural consistency of
M. genitalium protein. However, a range of M. genitalium variants still requires a structural
and functional assessment to evaluate their biological potential. The present findings from the
existing bioinformatics tool despite their accuracy might not be generalized across the wider
genome pool of M. genitalium because of their limited scalability. However, the study has
substantially enhanced the insight regarding the biological structure and function of the
presented M. genitalium protein sequence.
Introduction
The genital mycoplasmas categorize Mycoplasma genitalium as a microbe that leads to the
development of sexually transmitted infections in the human population (Sethi, Singh,
Samanta, & Sharma, 2012). Evidence-based literature describes the flask-shaped morphology
of Mycoplasma genitalium based on its terminal orgallene (i.e. cell membrane protrusion).
This organelle is a virulence factor that facilitates the motility and adhesion of Mycoplasma
genitalium across the host cell line. The cytoskeleton of the terminal organelle of
Mycoplasma genitalium corresponds to 300nm and incorporates the wheel complex, rod, and
terminal button (Martinelli et al., 2016). The crystal structure of Mycoplasma genitalium
protein (MG491) is based on a resolution of 3-angstrom. The subunits of Mycoplasma
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

P a g e | 2
genitalium protein incorporate the 60-residue N-terminus that includes the flexible C-terminal
location based on 150 residues of a centralized 3-helix bundle. This location communicates
with a distantly located terminal organelle protein (i.e. MG200). The MG491 protein of
Mycoplasma genitalium contains symmetrically oriented dimer subunits that configure the
shape of a tetramer. The genome of Mycoplasma genitalium is based on 580kb genome with
a small structure (Iverson-Cabral, Astete, Cohen, Rocha, & Totten, 2006). The MgPar
sequences replicate repeatedly in Mycoplasma genitalium in relation to their shared ancestry
with the mgpB gene. MgPar and mgpB sequences undergo recombination episodes to
facilitate the configuration of MgPa proteins. The whole genome sequencing of Mycoplasma
genitalium by Fookes et al. (2017) revealed twenty-five locations that exhibited elevated SNP
density levels. These locations were based on the host interactions of MgPar loci. The
phylogenetic assessment of Mycoplasma genitalium revealed the existence of 2-distinct clads
with variable genetic structure. The SNP locations including parC and 23S rRNA (V-regions)
of Mycoplasma genitalium evidently contributed to its resistance against fluoroquinolone,
erythromycin, and azithromycin (Fookes et al., 2017). These SNPs were also found to be
consistent with minimum inhibitory concentration data of the respective microorganisms. The
proteome/complete genome assessment of Mycoplasma genitalium by Ma, et al. (2007)
revealed the recombination episodes through gene conversion/crossover between MgPar
sequences and MG192 expression location that resulted in several variations in the MG192
sequence.
The evidence-based findings reveal several sequence combinations/alliances that lead to the
significant structural/functional variability within the proteins of Mycoplasma genitalium. A
range of significant bioinformatics-based databases assists in evaluating the structure of
complex proteins. Some of the significant databases include UniProtKB, UniGene, GenBank,
and RefSeq (NCBI Reference Sequence Database) (Chen, Huang, & Wu, 2017). The
significant protein search tools include ‘BLAST’, ‘LinkOut’, ‘E-Utilities’, and ‘Batch
Entrez’. PROSITE assists in evaluating the functional patterns, locations, and domains of
significant proteins (PROSITE, 2019). Alternatively, the tools including Pfam, PRINTS,
ProDOM, Blocks, and MOTIF substantially assist in analyzing various protein sequences in
accordance with their multifactorial parameters (MOTIFS, 2019). The presented paper
effectively analyses the preselected protein sequence pertaining to the hypothetical protein
‘Mycoplasma genitalium’ across various protein sequence databases. The paper evaluates the
domain structure, likely function, appearance, comparative appearance (in other
genitalium protein incorporate the 60-residue N-terminus that includes the flexible C-terminal
location based on 150 residues of a centralized 3-helix bundle. This location communicates
with a distantly located terminal organelle protein (i.e. MG200). The MG491 protein of
Mycoplasma genitalium contains symmetrically oriented dimer subunits that configure the
shape of a tetramer. The genome of Mycoplasma genitalium is based on 580kb genome with
a small structure (Iverson-Cabral, Astete, Cohen, Rocha, & Totten, 2006). The MgPar
sequences replicate repeatedly in Mycoplasma genitalium in relation to their shared ancestry
with the mgpB gene. MgPar and mgpB sequences undergo recombination episodes to
facilitate the configuration of MgPa proteins. The whole genome sequencing of Mycoplasma
genitalium by Fookes et al. (2017) revealed twenty-five locations that exhibited elevated SNP
density levels. These locations were based on the host interactions of MgPar loci. The
phylogenetic assessment of Mycoplasma genitalium revealed the existence of 2-distinct clads
with variable genetic structure. The SNP locations including parC and 23S rRNA (V-regions)
of Mycoplasma genitalium evidently contributed to its resistance against fluoroquinolone,
erythromycin, and azithromycin (Fookes et al., 2017). These SNPs were also found to be
consistent with minimum inhibitory concentration data of the respective microorganisms. The
proteome/complete genome assessment of Mycoplasma genitalium by Ma, et al. (2007)
revealed the recombination episodes through gene conversion/crossover between MgPar
sequences and MG192 expression location that resulted in several variations in the MG192
sequence.
The evidence-based findings reveal several sequence combinations/alliances that lead to the
significant structural/functional variability within the proteins of Mycoplasma genitalium. A
range of significant bioinformatics-based databases assists in evaluating the structure of
complex proteins. Some of the significant databases include UniProtKB, UniGene, GenBank,
and RefSeq (NCBI Reference Sequence Database) (Chen, Huang, & Wu, 2017). The
significant protein search tools include ‘BLAST’, ‘LinkOut’, ‘E-Utilities’, and ‘Batch
Entrez’. PROSITE assists in evaluating the functional patterns, locations, and domains of
significant proteins (PROSITE, 2019). Alternatively, the tools including Pfam, PRINTS,
ProDOM, Blocks, and MOTIF substantially assist in analyzing various protein sequences in
accordance with their multifactorial parameters (MOTIFS, 2019). The presented paper
effectively analyses the preselected protein sequence pertaining to the hypothetical protein
‘Mycoplasma genitalium’ across various protein sequence databases. The paper evaluates the
domain structure, likely function, appearance, comparative appearance (in other

P a g e | 3
organisms/life kingdoms), sequence alignment patterns, structure of related sequences,
transmembrane segments, and secondary structure of the selected protein sequence.
Methods
The presented study effectively evaluated the following protein sequence pertaining to M.
genitalium through the utilization of bioinformatics tool, techniques, methods, and databases.
MKKQLKYCFF SLFVSLSSIL SSCGSTTFVL ANFESYISPL LLERVQEKHP
LTFLTYPSNE
KLINGFANNT YSVAVASTYA VSELIERDLL SPIDWSQFNL KKSSSSSDKV
NNASDAKDLF
IDSIKEISQQ TKDSKNNELL HWAVPYFLQN LVFVYRGEKI SELEQENVSW
TDVIKAIVKH
KDRFNDNRLV FIDDARTIFS LANIVNTNNN SADVNPKEDG IGYFTNVYES
FQRLGLTKSN
LDSIFVNSDS NIVINELASG RRQGGIVYNG DAVYAALGGD LRDELSEEQI
PDGNNFHIVQ
PKISPVALDL LVINKQQSNF QKEAHEIIFD LALDGADQTK EQLIKTDEEL
GTDDEDFYLK
GAMQNFSYVN YVSPLKVISD PSTGIVSSKK NNAEMKSKQM STDQMTSEKE
FDYYTETLKA
LLEKEDSAEL NENEKKLVET IKKAYTIEKD SSIRWNQLVE KPISPLQRSN
LSLSWLDFKL
HWW
The study utilized Blast-p, PSI-BLAST, FASTA, Expasy, PROSITE, Pfam, EBI, and
SwissProt to evaluate and coherently explore/analyze the protein sequence outcomes and
structures. The BLAST findings in retrieving the significant M. genitalium sequences along
with their alignments. The extraction of gaps and identities assisted in predicting the
structural configuration of the retrieved protein variants. BLAST also assisted in evaluating
the identical-proteins and the comparatively analyzing different protein structures for
predicting their associated lipoproteins and functional variations. FASTA tool assisted in
evaluating the structural variants of Mycoplasma genitalium and PROSITE evaluation helped
in predicting the lipids and their structural/functional variations. The SWISS model proved to
be a significant tool in exploring the secondary protein structures for their comparative
assessment. This selected bioinformatics tools provided a significant interface to explore and
organisms/life kingdoms), sequence alignment patterns, structure of related sequences,
transmembrane segments, and secondary structure of the selected protein sequence.
Methods
The presented study effectively evaluated the following protein sequence pertaining to M.
genitalium through the utilization of bioinformatics tool, techniques, methods, and databases.
MKKQLKYCFF SLFVSLSSIL SSCGSTTFVL ANFESYISPL LLERVQEKHP
LTFLTYPSNE
KLINGFANNT YSVAVASTYA VSELIERDLL SPIDWSQFNL KKSSSSSDKV
NNASDAKDLF
IDSIKEISQQ TKDSKNNELL HWAVPYFLQN LVFVYRGEKI SELEQENVSW
TDVIKAIVKH
KDRFNDNRLV FIDDARTIFS LANIVNTNNN SADVNPKEDG IGYFTNVYES
FQRLGLTKSN
LDSIFVNSDS NIVINELASG RRQGGIVYNG DAVYAALGGD LRDELSEEQI
PDGNNFHIVQ
PKISPVALDL LVINKQQSNF QKEAHEIIFD LALDGADQTK EQLIKTDEEL
GTDDEDFYLK
GAMQNFSYVN YVSPLKVISD PSTGIVSSKK NNAEMKSKQM STDQMTSEKE
FDYYTETLKA
LLEKEDSAEL NENEKKLVET IKKAYTIEKD SSIRWNQLVE KPISPLQRSN
LSLSWLDFKL
HWW
The study utilized Blast-p, PSI-BLAST, FASTA, Expasy, PROSITE, Pfam, EBI, and
SwissProt to evaluate and coherently explore/analyze the protein sequence outcomes and
structures. The BLAST findings in retrieving the significant M. genitalium sequences along
with their alignments. The extraction of gaps and identities assisted in predicting the
structural configuration of the retrieved protein variants. BLAST also assisted in evaluating
the identical-proteins and the comparatively analyzing different protein structures for
predicting their associated lipoproteins and functional variations. FASTA tool assisted in
evaluating the structural variants of Mycoplasma genitalium and PROSITE evaluation helped
in predicting the lipids and their structural/functional variations. The SWISS model proved to
be a significant tool in exploring the secondary protein structures for their comparative
assessment. This selected bioinformatics tools provided a significant interface to explore and
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

P a g e | 4
align the protein variants in the context of correlating their structural
similarities/dissimilarities with the predicted functional outcomes. In summary, the selected
tools provided a platform to predict, model, analyze, integrate, and visualize the provided M.
genitalium sequence with a substantial matching score.
Results and Discussion
Sequence Analysis Through Blast-p and FASTA Algorithms (Structural and Functional
Assessment of the Conserved Putative Domains)
The analysis of the selected protein sequence through Blast-p tool revealed the occurrence of
putative conserved domains that included Lipoprotein_8, PotD, and PBP2_PotD_PotF_like
components (BLAST_Query_52229, 2019). Lipoprotein 8 revealed the hypothetical
lipoprotein that pertained to the solute binding family ‘MG045’. This finding suggests the
solute binding potential of Lipoprotein-8. PotD belonged to the periplasmic
Putrescine/Spermidine-binding protein associated with amino acid metabolism and transport.
The ABC transporters’ periplasmic-binding element was represented by
‘PBP2_PotD_PotF_like’ family that exhibited the capacity to facilitate polyamines' uptake.
This element included the periplasmic-binding fold (type II) (BLAST_Query_17409, 2019).
The family of this periplasmic substrate-binding conserved domain coordinates with
polyamine (ABC-type) transport processes in the form of elevated affinity primary receptors
that effectively regulate the intracellular levels of polyamine in various tumors and
proliferating cells. The polyamines including spermine, spermidine, and putrescine are the
natural polyamines that facilitate disease resistance, cellular proliferation/survival, and cell
growth. The natural polyamines also modulate the function of nucleic acids and other
negatively charged molecules. PBP2 family members represent periplasmic-binding (type II)
fold superfamily that contains the globular subdomains that connect with their individualized
ligands through the formation of a flexible hinge pattern. The binding of PBP2 ligands occurs
across the interdomain clefts while following a Venus flytrap pattern.
align the protein variants in the context of correlating their structural
similarities/dissimilarities with the predicted functional outcomes. In summary, the selected
tools provided a platform to predict, model, analyze, integrate, and visualize the provided M.
genitalium sequence with a substantial matching score.
Results and Discussion
Sequence Analysis Through Blast-p and FASTA Algorithms (Structural and Functional
Assessment of the Conserved Putative Domains)
The analysis of the selected protein sequence through Blast-p tool revealed the occurrence of
putative conserved domains that included Lipoprotein_8, PotD, and PBP2_PotD_PotF_like
components (BLAST_Query_52229, 2019). Lipoprotein 8 revealed the hypothetical
lipoprotein that pertained to the solute binding family ‘MG045’. This finding suggests the
solute binding potential of Lipoprotein-8. PotD belonged to the periplasmic
Putrescine/Spermidine-binding protein associated with amino acid metabolism and transport.
The ABC transporters’ periplasmic-binding element was represented by
‘PBP2_PotD_PotF_like’ family that exhibited the capacity to facilitate polyamines' uptake.
This element included the periplasmic-binding fold (type II) (BLAST_Query_17409, 2019).
The family of this periplasmic substrate-binding conserved domain coordinates with
polyamine (ABC-type) transport processes in the form of elevated affinity primary receptors
that effectively regulate the intracellular levels of polyamine in various tumors and
proliferating cells. The polyamines including spermine, spermidine, and putrescine are the
natural polyamines that facilitate disease resistance, cellular proliferation/survival, and cell
growth. The natural polyamines also modulate the function of nucleic acids and other
negatively charged molecules. PBP2 family members represent periplasmic-binding (type II)
fold superfamily that contains the globular subdomains that connect with their individualized
ligands through the formation of a flexible hinge pattern. The binding of PBP2 ligands occurs
across the interdomain clefts while following a Venus flytrap pattern.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
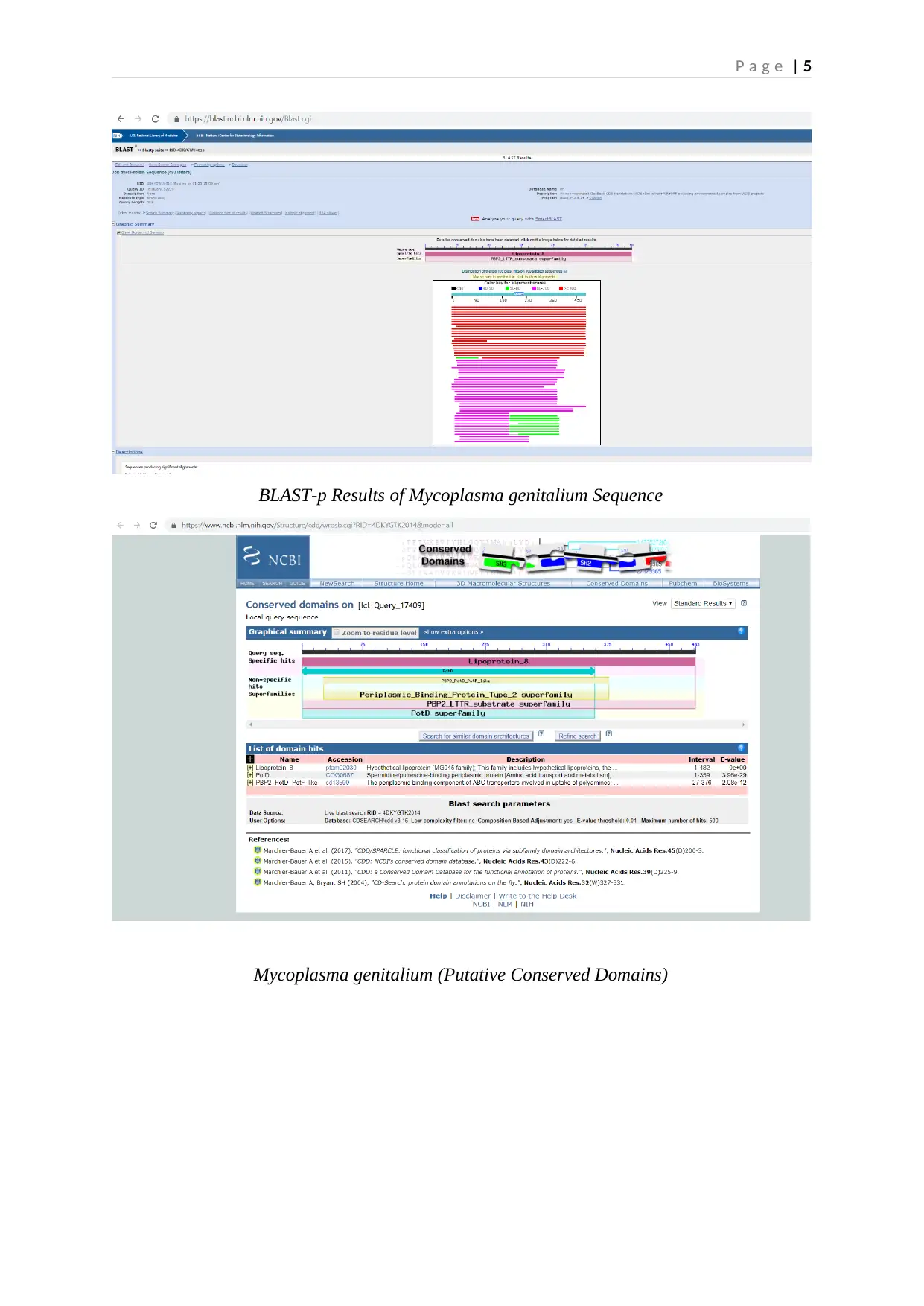
P a g e | 5
BLAST-p Results of Mycoplasma genitalium Sequence
Mycoplasma genitalium (Putative Conserved Domains)
BLAST-p Results of Mycoplasma genitalium Sequence
Mycoplasma genitalium (Putative Conserved Domains)

P a g e | 6
M. genitalium (Basic Details)
Domain Structure Assessment Through FASTA revealed the following output
(FASTA_Query_I20190122-125408-0265-15530702-p2m, 2019).
M. genitalium (Basic Details)
Domain Structure Assessment Through FASTA revealed the following output
(FASTA_Query_I20190122-125408-0265-15530702-p2m, 2019).
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

P a g e | 7
FASTA analysis revealed the structural variant of mycoplasma that incorporated MG045
(uncharacterized lipoprotein). The protein sequence revealed 483aa overlap, 100% identity,
and Smith-Waterman/Z-scores of 3099/3639.1 respectively. The bioactive molecules
revealed from the queried sequence included the palmitoyl group, MG045 (uncharacterized
lipoprotein), N-palmitoyl cysteine, S-diacylglycerol cysteine, cysteine zwitterion, diglyceride,
and L-cysteine. The sequence alignment of the uncharacterized lipoprotein (MG045)
obtained through FASTA is shown below (P47291_(Y045_MYCGE), 2019).
FASTA analysis revealed the structural variant of mycoplasma that incorporated MG045
(uncharacterized lipoprotein). The protein sequence revealed 483aa overlap, 100% identity,
and Smith-Waterman/Z-scores of 3099/3639.1 respectively. The bioactive molecules
revealed from the queried sequence included the palmitoyl group, MG045 (uncharacterized
lipoprotein), N-palmitoyl cysteine, S-diacylglycerol cysteine, cysteine zwitterion, diglyceride,
and L-cysteine. The sequence alignment of the uncharacterized lipoprotein (MG045)
obtained through FASTA is shown below (P47291_(Y045_MYCGE), 2019).
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

P a g e | 8
The taxonomic assessment of the Mycoplasma genitalium sequence through Uniprot revealed
its relationship/lineage with the cellular organisms. The organism is also known by the stains
including ATCC 33530 and G37. The findings also reveal the small structure of the retrieved
sequence as compared to other known organisms. These findings effectively concord with the
evidence-based literature. For example, the assessment of Mycoplasma genitalium genome’s
580070 base-pairs or complete nucleotide sequence by (Fraser et al., 1995) revealed its
smallest structure in comparison to other free-living organisms. The assessment also
suggested 470 coding locations that incorporate the genes warranted for energy metabolism,
cellular transport, DNA repair, translation, transcription, and DNA replication. The structural
and functional comparison of Mycoplasma genitalium genome with Haemophilus influenzae
genome revealed substantial variations between the metabolic capacity and physiology of
both microbes.
The partial sequencing of 508 randomized Mycoplasma genitalium clones by (Peterson, Hu,
Bott, & Hutchison, 1993) revealed 291 discrete contigs that constituted 17% genome of the
selected organism. The comparative analysis of 100993 nucleotide sequences in relation to
protein sequence databases and DNA revealed 46% homology to the known protein
sequences of the organism. The findings revealed multiple genes of the Mycoplasma
genitalium genome that facilitated significant metabolic systems and processes. These
findings closely match with the outcomes of the BLAST search algorithm of the selected M.
genitalium sequence that revealed 5 different types of hypothetical protein sequences
The taxonomic assessment of the Mycoplasma genitalium sequence through Uniprot revealed
its relationship/lineage with the cellular organisms. The organism is also known by the stains
including ATCC 33530 and G37. The findings also reveal the small structure of the retrieved
sequence as compared to other known organisms. These findings effectively concord with the
evidence-based literature. For example, the assessment of Mycoplasma genitalium genome’s
580070 base-pairs or complete nucleotide sequence by (Fraser et al., 1995) revealed its
smallest structure in comparison to other free-living organisms. The assessment also
suggested 470 coding locations that incorporate the genes warranted for energy metabolism,
cellular transport, DNA repair, translation, transcription, and DNA replication. The structural
and functional comparison of Mycoplasma genitalium genome with Haemophilus influenzae
genome revealed substantial variations between the metabolic capacity and physiology of
both microbes.
The partial sequencing of 508 randomized Mycoplasma genitalium clones by (Peterson, Hu,
Bott, & Hutchison, 1993) revealed 291 discrete contigs that constituted 17% genome of the
selected organism. The comparative analysis of 100993 nucleotide sequences in relation to
protein sequence databases and DNA revealed 46% homology to the known protein
sequences of the organism. The findings revealed multiple genes of the Mycoplasma
genitalium genome that facilitated significant metabolic systems and processes. These
findings closely match with the outcomes of the BLAST search algorithm of the selected M.
genitalium sequence that revealed 5 different types of hypothetical protein sequences

P a g e | 9
pertaining to different structure and functions. The evidence-based findings also reveal 4%
DNA repetitions and 390 proteins in M. genitalium genome. These findings matched with the
BLAST findings of M. genitalium genome sequence that revealed a total of 483 proteins
across the genome database. The presented sequence analysis, as well as evidence-based
literature, also revealed various cellular functions of M. genitalium genes.
Sequence Analysis Through PROSITE and Pfam
The PROSITE assessment of the M. genitalium sequence retrieved the following outcomes.
The sequence assessment revealed the lipids, including N-palmitoyl cysteine and S-
diacylglycerol cysteine. Evidence-based findings reveal the dynamic biological activity of
lipopeptides that embed free NH2 terminus with palmitoyl (ester-bounded) residues (You,
Zeng, & Wu, 2006). These lipopeptides substantially induce macrophages and exhibit greater
affinity for the murine splenocytes. These findings recommend the biological activity of M.
genitalium against the immune responses as well as the cell-mediated immunity. The
presence of S-diacylglycerol cysteine in M. gentalium sequence also affirms its affinity for
the biological pathways. This is evidenced by the capacity of S-diacylglycerol-L-cysteine
molecule to configure Bronsted acid or base through hydron acquisition or donation (ChEBI,
2014).
The Pfam assessment of the selected sequence revealed the following outcomes.
pertaining to different structure and functions. The evidence-based findings also reveal 4%
DNA repetitions and 390 proteins in M. genitalium genome. These findings matched with the
BLAST findings of M. genitalium genome sequence that revealed a total of 483 proteins
across the genome database. The presented sequence analysis, as well as evidence-based
literature, also revealed various cellular functions of M. genitalium genes.
Sequence Analysis Through PROSITE and Pfam
The PROSITE assessment of the M. genitalium sequence retrieved the following outcomes.
The sequence assessment revealed the lipids, including N-palmitoyl cysteine and S-
diacylglycerol cysteine. Evidence-based findings reveal the dynamic biological activity of
lipopeptides that embed free NH2 terminus with palmitoyl (ester-bounded) residues (You,
Zeng, & Wu, 2006). These lipopeptides substantially induce macrophages and exhibit greater
affinity for the murine splenocytes. These findings recommend the biological activity of M.
genitalium against the immune responses as well as the cell-mediated immunity. The
presence of S-diacylglycerol cysteine in M. gentalium sequence also affirms its affinity for
the biological pathways. This is evidenced by the capacity of S-diacylglycerol-L-cysteine
molecule to configure Bronsted acid or base through hydron acquisition or donation (ChEBI,
2014).
The Pfam assessment of the selected sequence revealed the following outcomes.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

P a g e | 10
The hypothetical protein retrieved through the Pfam search pertains to the MG045 family.
The lipoprotein-8 obtained through the Pfam tool is a hypothetical lipoprotein that
corresponds to solute binding proteins’ family (i.e. pfam01547). This evidence-based
outcome reveals the solute binding property of lipoprotein-8
(NCBI_pfam02030_Lipoprotein_8, 2017). This property is presumably exhibited by M.
genitalium while infecting the host cell lines.
Identical Proteins
Several identical lipoproteins of M. genitalium were obtained through the BlASTp search
algorithm (BLAST_IPG, 2019). One of the predicted lipoproteins correlated with HMM
PF02030 protein family and appeared to be a putrescine/spermidine/putative/permease ABC
transporter. The putrescine/spermidine binding/periplasmic proteins assist the polyamine
mechanism that facilitates the spermidine/putrescine transport across the bacterial periplasm
(InterPro, 2019). These properties of Mycoplasma genitalium sequence explain its pathogenic
potential inside the host cells.
The hypothetical protein retrieved through the Pfam search pertains to the MG045 family.
The lipoprotein-8 obtained through the Pfam tool is a hypothetical lipoprotein that
corresponds to solute binding proteins’ family (i.e. pfam01547). This evidence-based
outcome reveals the solute binding property of lipoprotein-8
(NCBI_pfam02030_Lipoprotein_8, 2017). This property is presumably exhibited by M.
genitalium while infecting the host cell lines.
Identical Proteins
Several identical lipoproteins of M. genitalium were obtained through the BlASTp search
algorithm (BLAST_IPG, 2019). One of the predicted lipoproteins correlated with HMM
PF02030 protein family and appeared to be a putrescine/spermidine/putative/permease ABC
transporter. The putrescine/spermidine binding/periplasmic proteins assist the polyamine
mechanism that facilitates the spermidine/putrescine transport across the bacterial periplasm
(InterPro, 2019). These properties of Mycoplasma genitalium sequence explain its pathogenic
potential inside the host cells.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
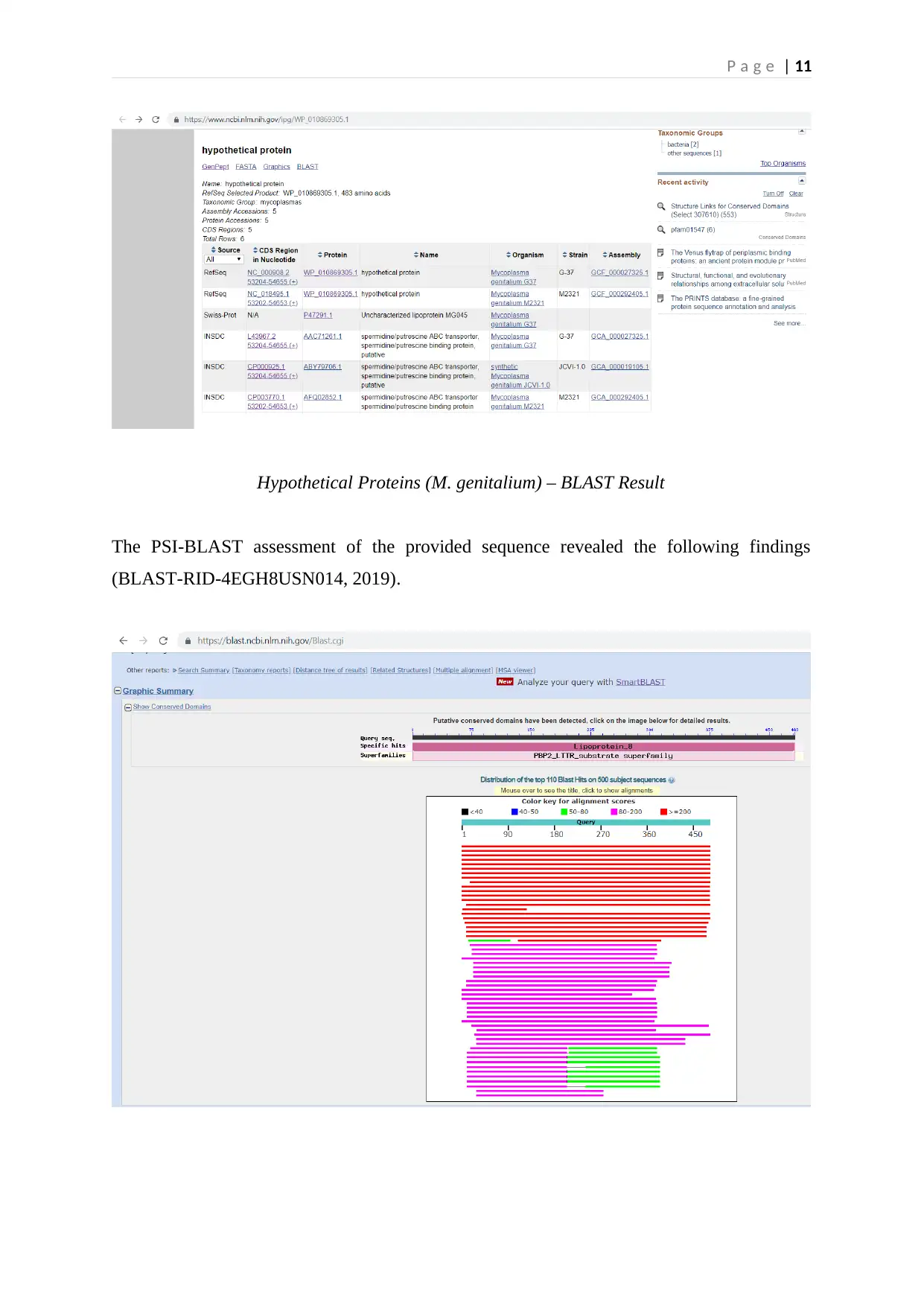
P a g e | 11
Hypothetical Proteins (M. genitalium) – BLAST Result
The PSI-BLAST assessment of the provided sequence revealed the following findings
(BLAST-RID-4EGH8USN014, 2019).
Hypothetical Proteins (M. genitalium) – BLAST Result
The PSI-BLAST assessment of the provided sequence revealed the following findings
(BLAST-RID-4EGH8USN014, 2019).

P a g e | 12
The molecular weight of the best matching sequence exhibited a molecular weight of 54745.
The retrieved hypothetical protein pertained to pfam02030 and MG045 family. The
periplasmic lipoprotein of the retrieved M. genitalium sequence pertained to the
putrescine/spermidine-binding location. The following identical sequences were also
obtained through the PSI-BLAST search.
No difference was recorded between the structures of the hypothetical proteins obtained
through BLAST-p and PSI-BLAST searches.
The molecular weight of the best matching sequence exhibited a molecular weight of 54745.
The retrieved hypothetical protein pertained to pfam02030 and MG045 family. The
periplasmic lipoprotein of the retrieved M. genitalium sequence pertained to the
putrescine/spermidine-binding location. The following identical sequences were also
obtained through the PSI-BLAST search.
No difference was recorded between the structures of the hypothetical proteins obtained
through BLAST-p and PSI-BLAST searches.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 22
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.