Protein Discovery and Analysis: Beta Glucuronidase Analysis Report
VerifiedAdded on 2022/07/27
|14
|2038
|50
Report
AI Summary
This report provides a comprehensive analysis of Beta Glucuronidase, a type of glucuronidase that catalyzes the hydrolysis of beta-D-glucuronic acid residues. The report begins by detailing the primary structure of the protein, including its amino acid sequence and highlighting glycosylation sites. It then explores the physico-chemical properties of the protein, such as molecular weight, theoretical pI, and amino acid composition, along with a hydropathy plot that visualizes the protein's hydrophobic and hydrophilic regions. The report also covers both initial and present purification methods, including immunoadsorption, DEAE-Sephadex chromatography, and recombination techniques, providing a detailed overview of the processes involved in isolating and purifying the protein. References to relevant research papers are included, providing context and supporting the findings presented in the report. Overall, the report offers a detailed examination of the structure, properties, and purification of Beta Glucuronidase.

Running head: PROTEIN DISCOVERY AND ANALYSIS
BETA GLUCORINIDASE
Name of the Student
Name of the University
Author Note
BETA GLUCORINIDASE
Name of the Student
Name of the University
Author Note
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

1PROTEIN DISCOVERY AND ANALYSIS
Table of Contents
Introduction................................................................................................................................2
The primary structure of PET protein........................................................................................2
The physico-chemical properties of PET protein.......................................................................3
Initial purification of the protein................................................................................................8
Present purification of the protein..............................................................................................9
Bibliography.............................................................................................................................11
Table of Contents
Introduction................................................................................................................................2
The primary structure of PET protein........................................................................................2
The physico-chemical properties of PET protein.......................................................................3
Initial purification of the protein................................................................................................8
Present purification of the protein..............................................................................................9
Bibliography.............................................................................................................................11

2PROTEIN DISCOVERY AND ANALYSIS
Introduction
Human beta glucuronidase has been defied as a type of glucuronidase which catalyzes the
hydrolysis of beta D glucuronic acid residue that has been found derived from a non-reducing
mutopolysaccharide in nature. This protein has been found to be located in the lysozyme.
Beta glucuronidase has been found to be secreted from gusB gene of human (Di et al., 2017).
The protein is synthesised in the form of a monomer of 80 kDa size before proteolysis. After
proteolysis, it has been found to be broken down to 78 kDa monomer, which is the active
protein.
The primary structure of PET protein
10 20 30 40 50
MARGSAVAWA ALGPLLWGCA LGLQGGMLYP QESPSRECKE LDGLWSFRAD
60 70 80 90 100
FSDNRRRGFE EQWYRRPLWE SGPTVDMPVP SSFNDISQDW RLRHFVGWVW
110 120 130 140 150
YEREVILPER WTQDLRTRVV LRIGSAHSYA IVWVNGVDTL EHEGGYLPFE
160 170 180 190 200
ADISNLVQVG PLPSRLRITI AINNTLTPTT LPPGTIQYLT DTSKYPKGYF
210 220 230 240 250
VQNTYFDFFN YAGLQRSVLL YTTPTTYIDD ITVTTSVEQD SGLVNYQISV
260 270 280 290 300
KGSNLFKLEV RLLDAENKVV ANGTGTQGQL KVPGVSLWWP YLMHERPAYL
310 320 330 340 350
YSLEVQLTAQ TSLGPVSDFY TLPVGIRTVA VTKSQFLING KPFYFHGVNK
360 370 380 390 400
HEDADIRGKG FDWPLLVKDF NLLRWLGANA FRTSHYPYAE EVMQMCDRYG
410 420 430 440 450
IVVIDECPGV GLALPQFFNN VSLHHHMQVM EEVVRRDKNH PAVVMWSVAN
460 470 480 490 500
EPASHLESAG YYLKMVIAHT KSLDPSRPVT FVSNSNYAAD KGAPYVDVIC
510 520 530 540 550
LNSYYSWYHD YGHLELIQLQ LATQFENWYK KYQKPIIQSE YGAETIAGFH
560 570 580 590 600
QDPPLMFTEE YQKSLLEQYH LGLDQKRRKY VVGELIWNFA DFMTEQSPTR
610 620 630 640 650
VLGNKKGIFT RQRQPKSAAF LLRERYWKIA NETRYPHSVA KSQCLENSLF
T
Introduction
Human beta glucuronidase has been defied as a type of glucuronidase which catalyzes the
hydrolysis of beta D glucuronic acid residue that has been found derived from a non-reducing
mutopolysaccharide in nature. This protein has been found to be located in the lysozyme.
Beta glucuronidase has been found to be secreted from gusB gene of human (Di et al., 2017).
The protein is synthesised in the form of a monomer of 80 kDa size before proteolysis. After
proteolysis, it has been found to be broken down to 78 kDa monomer, which is the active
protein.
The primary structure of PET protein
10 20 30 40 50
MARGSAVAWA ALGPLLWGCA LGLQGGMLYP QESPSRECKE LDGLWSFRAD
60 70 80 90 100
FSDNRRRGFE EQWYRRPLWE SGPTVDMPVP SSFNDISQDW RLRHFVGWVW
110 120 130 140 150
YEREVILPER WTQDLRTRVV LRIGSAHSYA IVWVNGVDTL EHEGGYLPFE
160 170 180 190 200
ADISNLVQVG PLPSRLRITI AINNTLTPTT LPPGTIQYLT DTSKYPKGYF
210 220 230 240 250
VQNTYFDFFN YAGLQRSVLL YTTPTTYIDD ITVTTSVEQD SGLVNYQISV
260 270 280 290 300
KGSNLFKLEV RLLDAENKVV ANGTGTQGQL KVPGVSLWWP YLMHERPAYL
310 320 330 340 350
YSLEVQLTAQ TSLGPVSDFY TLPVGIRTVA VTKSQFLING KPFYFHGVNK
360 370 380 390 400
HEDADIRGKG FDWPLLVKDF NLLRWLGANA FRTSHYPYAE EVMQMCDRYG
410 420 430 440 450
IVVIDECPGV GLALPQFFNN VSLHHHMQVM EEVVRRDKNH PAVVMWSVAN
460 470 480 490 500
EPASHLESAG YYLKMVIAHT KSLDPSRPVT FVSNSNYAAD KGAPYVDVIC
510 520 530 540 550
LNSYYSWYHD YGHLELIQLQ LATQFENWYK KYQKPIIQSE YGAETIAGFH
560 570 580 590 600
QDPPLMFTEE YQKSLLEQYH LGLDQKRRKY VVGELIWNFA DFMTEQSPTR
610 620 630 640 650
VLGNKKGIFT RQRQPKSAAF LLRERYWKIA NETRYPHSVA KSQCLENSLF
T
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

3PROTEIN DISCOVERY AND ANALYSIS
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
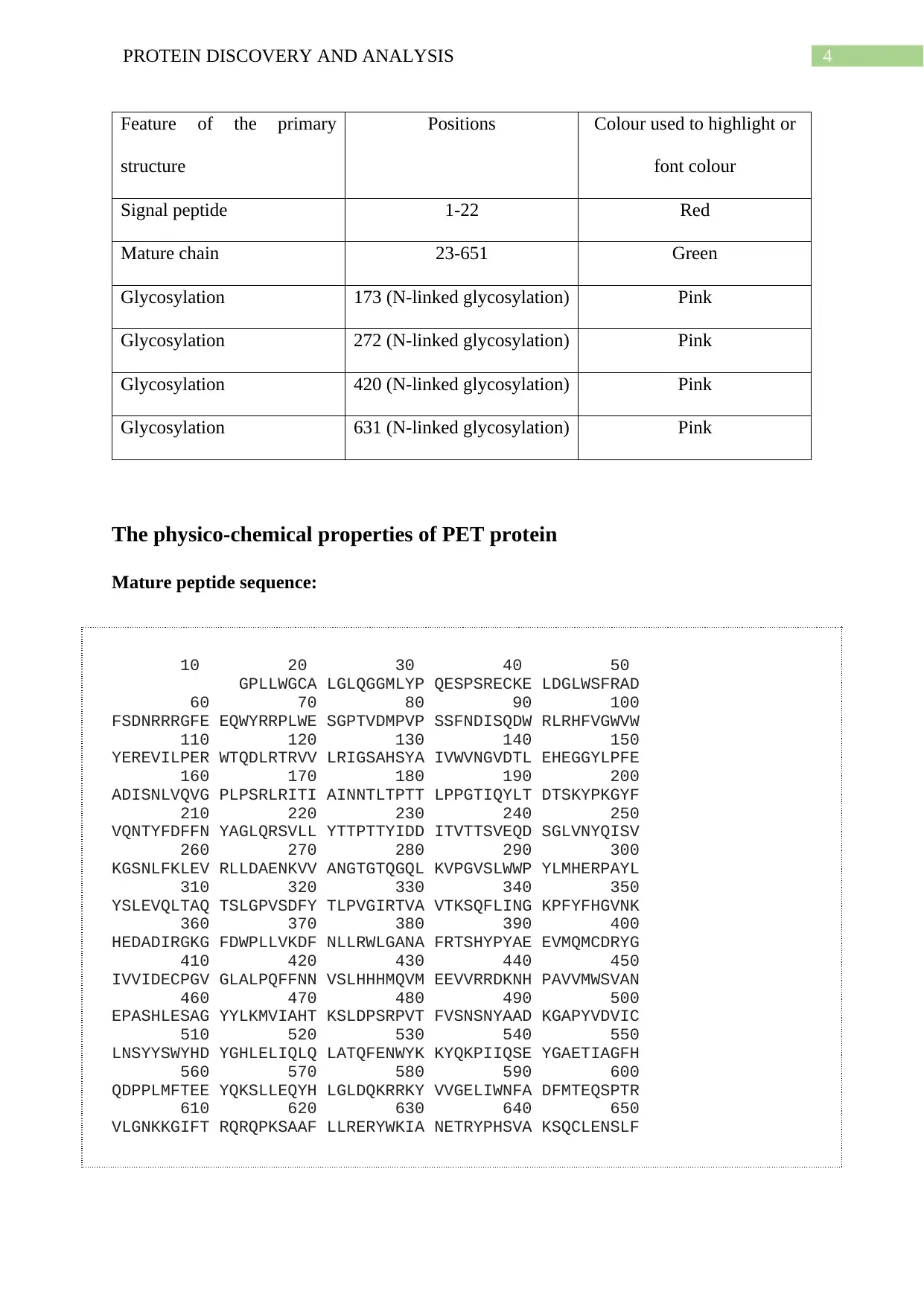
4PROTEIN DISCOVERY AND ANALYSIS
Feature of the primary
structure
Positions Colour used to highlight or
font colour
Signal peptide 1-22 Red
Mature chain 23-651 Green
Glycosylation 173 (N-linked glycosylation) Pink
Glycosylation 272 (N-linked glycosylation) Pink
Glycosylation 420 (N-linked glycosylation) Pink
Glycosylation 631 (N-linked glycosylation) Pink
The physico-chemical properties of PET protein
Mature peptide sequence:
10 20 30 40 50
GPLLWGCA LGLQGGMLYP QESPSRECKE LDGLWSFRAD
60 70 80 90 100
FSDNRRRGFE EQWYRRPLWE SGPTVDMPVP SSFNDISQDW RLRHFVGWVW
110 120 130 140 150
YEREVILPER WTQDLRTRVV LRIGSAHSYA IVWVNGVDTL EHEGGYLPFE
160 170 180 190 200
ADISNLVQVG PLPSRLRITI AINNTLTPTT LPPGTIQYLT DTSKYPKGYF
210 220 230 240 250
VQNTYFDFFN YAGLQRSVLL YTTPTTYIDD ITVTTSVEQD SGLVNYQISV
260 270 280 290 300
KGSNLFKLEV RLLDAENKVV ANGTGTQGQL KVPGVSLWWP YLMHERPAYL
310 320 330 340 350
YSLEVQLTAQ TSLGPVSDFY TLPVGIRTVA VTKSQFLING KPFYFHGVNK
360 370 380 390 400
HEDADIRGKG FDWPLLVKDF NLLRWLGANA FRTSHYPYAE EVMQMCDRYG
410 420 430 440 450
IVVIDECPGV GLALPQFFNN VSLHHHMQVM EEVVRRDKNH PAVVMWSVAN
460 470 480 490 500
EPASHLESAG YYLKMVIAHT KSLDPSRPVT FVSNSNYAAD KGAPYVDVIC
510 520 530 540 550
LNSYYSWYHD YGHLELIQLQ LATQFENWYK KYQKPIIQSE YGAETIAGFH
560 570 580 590 600
QDPPLMFTEE YQKSLLEQYH LGLDQKRRKY VVGELIWNFA DFMTEQSPTR
610 620 630 640 650
VLGNKKGIFT RQRQPKSAAF LLRERYWKIA NETRYPHSVA KSQCLENSLF
Feature of the primary
structure
Positions Colour used to highlight or
font colour
Signal peptide 1-22 Red
Mature chain 23-651 Green
Glycosylation 173 (N-linked glycosylation) Pink
Glycosylation 272 (N-linked glycosylation) Pink
Glycosylation 420 (N-linked glycosylation) Pink
Glycosylation 631 (N-linked glycosylation) Pink
The physico-chemical properties of PET protein
Mature peptide sequence:
10 20 30 40 50
GPLLWGCA LGLQGGMLYP QESPSRECKE LDGLWSFRAD
60 70 80 90 100
FSDNRRRGFE EQWYRRPLWE SGPTVDMPVP SSFNDISQDW RLRHFVGWVW
110 120 130 140 150
YEREVILPER WTQDLRTRVV LRIGSAHSYA IVWVNGVDTL EHEGGYLPFE
160 170 180 190 200
ADISNLVQVG PLPSRLRITI AINNTLTPTT LPPGTIQYLT DTSKYPKGYF
210 220 230 240 250
VQNTYFDFFN YAGLQRSVLL YTTPTTYIDD ITVTTSVEQD SGLVNYQISV
260 270 280 290 300
KGSNLFKLEV RLLDAENKVV ANGTGTQGQL KVPGVSLWWP YLMHERPAYL
310 320 330 340 350
YSLEVQLTAQ TSLGPVSDFY TLPVGIRTVA VTKSQFLING KPFYFHGVNK
360 370 380 390 400
HEDADIRGKG FDWPLLVKDF NLLRWLGANA FRTSHYPYAE EVMQMCDRYG
410 420 430 440 450
IVVIDECPGV GLALPQFFNN VSLHHHMQVM EEVVRRDKNH PAVVMWSVAN
460 470 480 490 500
EPASHLESAG YYLKMVIAHT KSLDPSRPVT FVSNSNYAAD KGAPYVDVIC
510 520 530 540 550
LNSYYSWYHD YGHLELIQLQ LATQFENWYK KYQKPIIQSE YGAETIAGFH
560 570 580 590 600
QDPPLMFTEE YQKSLLEQYH LGLDQKRRKY VVGELIWNFA DFMTEQSPTR
610 620 630 640 650
VLGNKKGIFT RQRQPKSAAF LLRERYWKIA NETRYPHSVA KSQCLENSLF

5PROTEIN DISCOVERY AND ANALYSIS
T
Physico-chemical properties:
User-provided sequence:
10 20 30 40 50 60
MARGSAVAWA ALGPLLWGCA LGLQGGMLYP QESPSRECKE LDGLWSFRAD FSDNRRRGFE
70 80 90 100 110 120
EQWYRRPLWE SGPTVDMPVP SSFNDISQDW RLRHFVGWVW YEREVILPER WTQDLRTRVV
130 140 150 160 170 180
LRIGSAHSYA IVWVNGVDTL EHEGGYLPFE ADISNLVQVG PLPSRLRITI AINNTLTPTT
190 200 210 220 230 240
LPPGTIQYLT DTSKYPKGYF VQNTYFDFFN YAGLQRSVLL YTTPTTYIDD ITVTTSVEQD
250 260 270 280 290 300
SGLVNYQISV KGSNLFKLEV RLLDAENKVV ANGTGTQGQL KVPGVSLWWP YLMHERPAYL
310 320 330 340 350 360
YSLEVQLTAQ TSLGPVSDFY TLPVGIRTVA VTKSQFLING KPFYFHGVNK HEDADIRGKG
370 380 390 400 410 420
FDWPLLVKDF NLLRWLGANA FRTSHYPYAE EVMQMCDRYG IVVIDECPGV GLALPQFFNN
430 440 450 460 470 480
VSLHHHMQVM EEVVRRDKNH PAVVMWSVAN EPASHLESAG YYLKMVIAHT KSLDPSRPVT
490 500 510 520 530 540
FVSNSNYAAD KGAPYVDVIC LNSYYSWYHD YGHLELIQLQ LATQFENWYK KYQKPIIQSE
550 560 570 580 590 600
YGAETIAGFH QDPPLMFTEE YQKSLLEQYH LGLDQKRRKY VVGELIWNFA DFMTEQSPTR
610 620 630 640 650
VLGNKKGIFT RQRQPKSAAF LLRERYWKIA NETRYPHSVA KSQCLENSLF T
Number of amino acids: 651
Molecular weight: 74731.94
Theoretical pI: 6.54
T
Physico-chemical properties:
User-provided sequence:
10 20 30 40 50 60
MARGSAVAWA ALGPLLWGCA LGLQGGMLYP QESPSRECKE LDGLWSFRAD FSDNRRRGFE
70 80 90 100 110 120
EQWYRRPLWE SGPTVDMPVP SSFNDISQDW RLRHFVGWVW YEREVILPER WTQDLRTRVV
130 140 150 160 170 180
LRIGSAHSYA IVWVNGVDTL EHEGGYLPFE ADISNLVQVG PLPSRLRITI AINNTLTPTT
190 200 210 220 230 240
LPPGTIQYLT DTSKYPKGYF VQNTYFDFFN YAGLQRSVLL YTTPTTYIDD ITVTTSVEQD
250 260 270 280 290 300
SGLVNYQISV KGSNLFKLEV RLLDAENKVV ANGTGTQGQL KVPGVSLWWP YLMHERPAYL
310 320 330 340 350 360
YSLEVQLTAQ TSLGPVSDFY TLPVGIRTVA VTKSQFLING KPFYFHGVNK HEDADIRGKG
370 380 390 400 410 420
FDWPLLVKDF NLLRWLGANA FRTSHYPYAE EVMQMCDRYG IVVIDECPGV GLALPQFFNN
430 440 450 460 470 480
VSLHHHMQVM EEVVRRDKNH PAVVMWSVAN EPASHLESAG YYLKMVIAHT KSLDPSRPVT
490 500 510 520 530 540
FVSNSNYAAD KGAPYVDVIC LNSYYSWYHD YGHLELIQLQ LATQFENWYK KYQKPIIQSE
550 560 570 580 590 600
YGAETIAGFH QDPPLMFTEE YQKSLLEQYH LGLDQKRRKY VVGELIWNFA DFMTEQSPTR
610 620 630 640 650
VLGNKKGIFT RQRQPKSAAF LLRERYWKIA NETRYPHSVA KSQCLENSLF T
Number of amino acids: 651
Molecular weight: 74731.94
Theoretical pI: 6.54
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

6PROTEIN DISCOVERY AND ANALYSIS
Amino acid composition obtained:
Ala (A) 38 5.8%
Arg (R) 36 5.5%
Asn (N) 28 4.3%
Asp (D) 30 4.6%
Cys (C) 6 0.9%
Gln (Q) 32 4.9%
Glu (E) 37 5.7%
Gly (G) 43 6.6%
His (H) 18 2.8%
Ile (I) 26 4.0%
Leu (L) 65 10.0%
Lys (K) 27 4.1%
Met (M) 12 1.8%
Phe (F) 29 4.5%
Pro (P) 37 5.7%
Ser (S) 42 6.5%
Thr (T) 38 5.8%
Trp (W) 19 2.9%
Tyr (Y) 37 5.7%
Val (V) 51 7.8%
Pyl (O) 0 0.0%
Sec (U) 0 0.0%
(B) 0 0.0%
(Z) 0 0.0%
(X) 0 0.0%
Total number of negatively charged residues (Asp + Glu): 67
Total number of positively charged residues (Arg + Lys): 63
Atomic composition:
Carbon C 3408
Amino acid composition obtained:
Ala (A) 38 5.8%
Arg (R) 36 5.5%
Asn (N) 28 4.3%
Asp (D) 30 4.6%
Cys (C) 6 0.9%
Gln (Q) 32 4.9%
Glu (E) 37 5.7%
Gly (G) 43 6.6%
His (H) 18 2.8%
Ile (I) 26 4.0%
Leu (L) 65 10.0%
Lys (K) 27 4.1%
Met (M) 12 1.8%
Phe (F) 29 4.5%
Pro (P) 37 5.7%
Ser (S) 42 6.5%
Thr (T) 38 5.8%
Trp (W) 19 2.9%
Tyr (Y) 37 5.7%
Val (V) 51 7.8%
Pyl (O) 0 0.0%
Sec (U) 0 0.0%
(B) 0 0.0%
(Z) 0 0.0%
(X) 0 0.0%
Total number of negatively charged residues (Asp + Glu): 67
Total number of positively charged residues (Arg + Lys): 63
Atomic composition:
Carbon C 3408
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
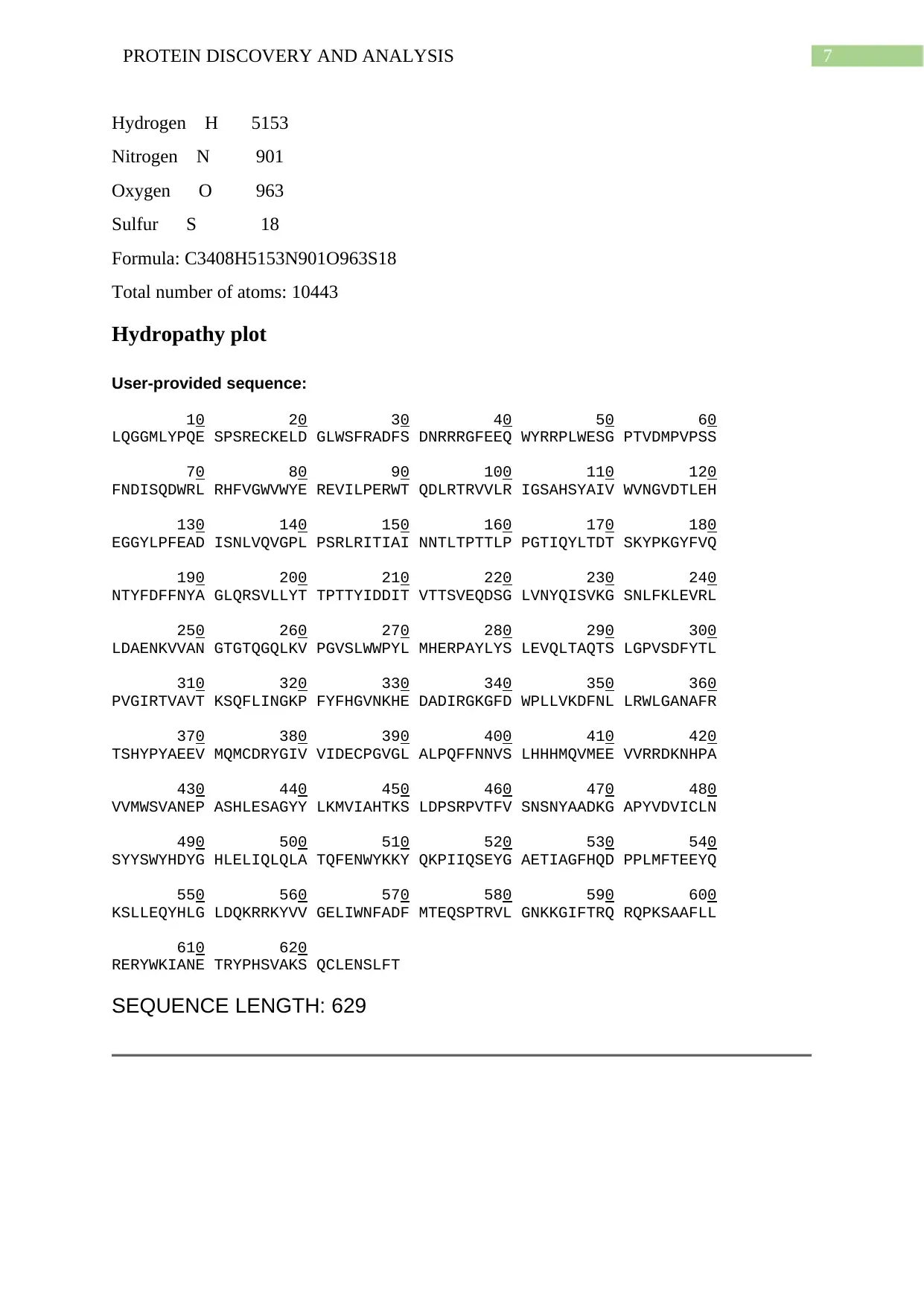
7PROTEIN DISCOVERY AND ANALYSIS
Hydrogen H 5153
Nitrogen N 901
Oxygen O 963
Sulfur S 18
Formula: C3408H5153N901O963S18
Total number of atoms: 10443
Hydropathy plot
User-provided sequence:
10 20 30 40 50 60
LQGGMLYPQE SPSRECKELD GLWSFRADFS DNRRRGFEEQ WYRRPLWESG PTVDMPVPSS
70 80 90 100 110 120
FNDISQDWRL RHFVGWVWYE REVILPERWT QDLRTRVVLR IGSAHSYAIV WVNGVDTLEH
130 140 150 160 170 180
EGGYLPFEAD ISNLVQVGPL PSRLRITIAI NNTLTPTTLP PGTIQYLTDT SKYPKGYFVQ
190 200 210 220 230 240
NTYFDFFNYA GLQRSVLLYT TPTTYIDDIT VTTSVEQDSG LVNYQISVKG SNLFKLEVRL
250 260 270 280 290 300
LDAENKVVAN GTGTQGQLKV PGVSLWWPYL MHERPAYLYS LEVQLTAQTS LGPVSDFYTL
310 320 330 340 350 360
PVGIRTVAVT KSQFLINGKP FYFHGVNKHE DADIRGKGFD WPLLVKDFNL LRWLGANAFR
370 380 390 400 410 420
TSHYPYAEEV MQMCDRYGIV VIDECPGVGL ALPQFFNNVS LHHHMQVMEE VVRRDKNHPA
430 440 450 460 470 480
VVMWSVANEP ASHLESAGYY LKMVIAHTKS LDPSRPVTFV SNSNYAADKG APYVDVICLN
490 500 510 520 530 540
SYYSWYHDYG HLELIQLQLA TQFENWYKKY QKPIIQSEYG AETIAGFHQD PPLMFTEEYQ
550 560 570 580 590 600
KSLLEQYHLG LDQKRRKYVV GELIWNFADF MTEQSPTRVL GNKKGIFTRQ RQPKSAAFLL
610 620
RERYWKIANE TRYPHSVAKS QCLENSLFT
SEQUENCE LENGTH: 629
Hydrogen H 5153
Nitrogen N 901
Oxygen O 963
Sulfur S 18
Formula: C3408H5153N901O963S18
Total number of atoms: 10443
Hydropathy plot
User-provided sequence:
10 20 30 40 50 60
LQGGMLYPQE SPSRECKELD GLWSFRADFS DNRRRGFEEQ WYRRPLWESG PTVDMPVPSS
70 80 90 100 110 120
FNDISQDWRL RHFVGWVWYE REVILPERWT QDLRTRVVLR IGSAHSYAIV WVNGVDTLEH
130 140 150 160 170 180
EGGYLPFEAD ISNLVQVGPL PSRLRITIAI NNTLTPTTLP PGTIQYLTDT SKYPKGYFVQ
190 200 210 220 230 240
NTYFDFFNYA GLQRSVLLYT TPTTYIDDIT VTTSVEQDSG LVNYQISVKG SNLFKLEVRL
250 260 270 280 290 300
LDAENKVVAN GTGTQGQLKV PGVSLWWPYL MHERPAYLYS LEVQLTAQTS LGPVSDFYTL
310 320 330 340 350 360
PVGIRTVAVT KSQFLINGKP FYFHGVNKHE DADIRGKGFD WPLLVKDFNL LRWLGANAFR
370 380 390 400 410 420
TSHYPYAEEV MQMCDRYGIV VIDECPGVGL ALPQFFNNVS LHHHMQVMEE VVRRDKNHPA
430 440 450 460 470 480
VVMWSVANEP ASHLESAGYY LKMVIAHTKS LDPSRPVTFV SNSNYAADKG APYVDVICLN
490 500 510 520 530 540
SYYSWYHDYG HLELIQLQLA TQFENWYKKY QKPIIQSEYG AETIAGFHQD PPLMFTEEYQ
550 560 570 580 590 600
KSLLEQYHLG LDQKRRKYVV GELIWNFADF MTEQSPTRVL GNKKGIFTRQ RQPKSAAFLL
610 620
RERYWKIANE TRYPHSVAKS QCLENSLFT
SEQUENCE LENGTH: 629
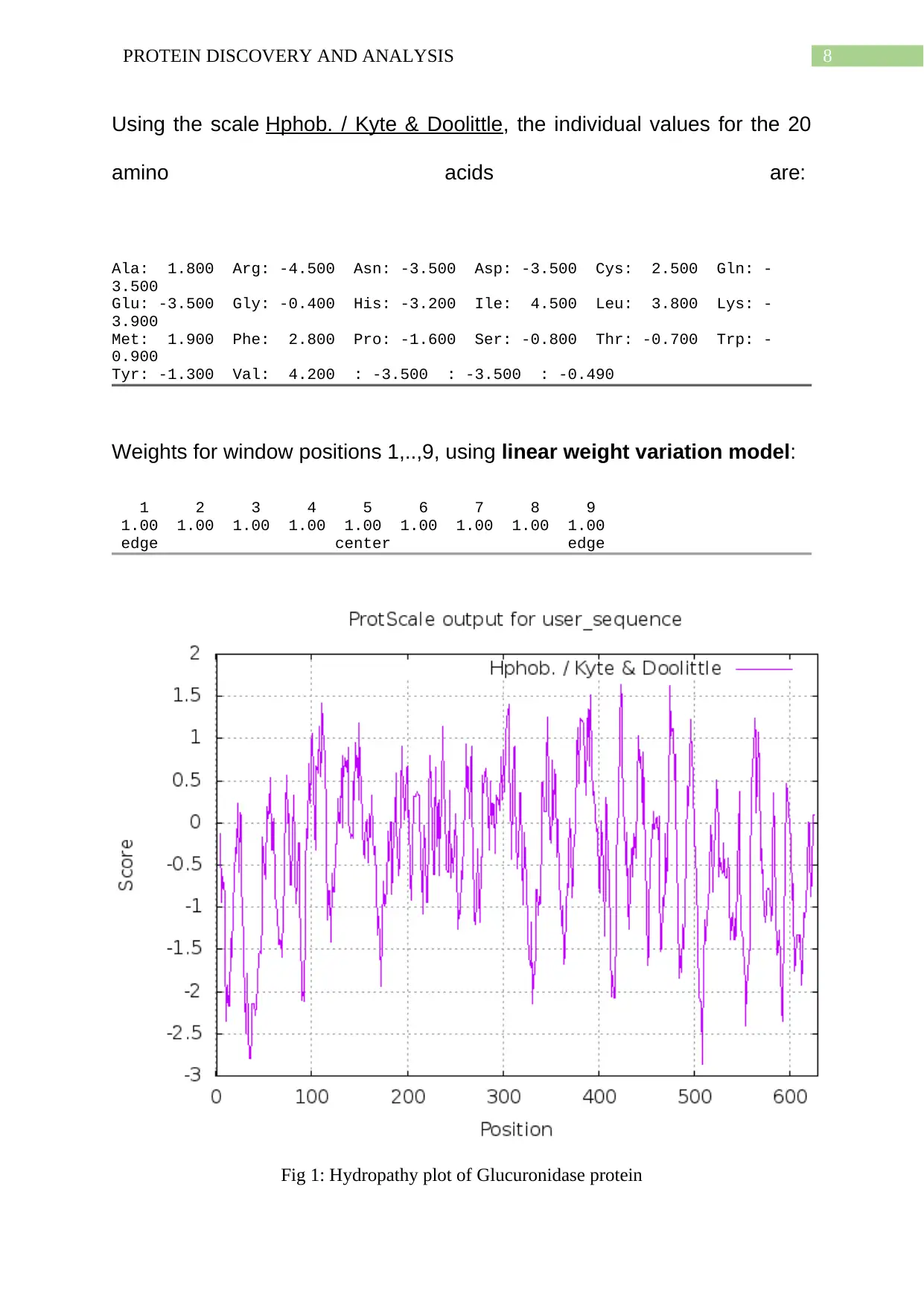
8PROTEIN DISCOVERY AND ANALYSIS
Using the scale Hphob. / Kyte & Doolittle, the individual values for the 20
amino acids are:
Ala: 1.800 Arg: -4.500 Asn: -3.500 Asp: -3.500 Cys: 2.500 Gln: -
3.500
Glu: -3.500 Gly: -0.400 His: -3.200 Ile: 4.500 Leu: 3.800 Lys: -
3.900
Met: 1.900 Phe: 2.800 Pro: -1.600 Ser: -0.800 Thr: -0.700 Trp: -
0.900
Tyr: -1.300 Val: 4.200 : -3.500 : -3.500 : -0.490
Weights for window positions 1,..,9, using linear weight variation model:
1 2 3 4 5 6 7 8 9
1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00
edge center edge
Fig 1: Hydropathy plot of Glucuronidase protein
Using the scale Hphob. / Kyte & Doolittle, the individual values for the 20
amino acids are:
Ala: 1.800 Arg: -4.500 Asn: -3.500 Asp: -3.500 Cys: 2.500 Gln: -
3.500
Glu: -3.500 Gly: -0.400 His: -3.200 Ile: 4.500 Leu: 3.800 Lys: -
3.900
Met: 1.900 Phe: 2.800 Pro: -1.600 Ser: -0.800 Thr: -0.700 Trp: -
0.900
Tyr: -1.300 Val: 4.200 : -3.500 : -3.500 : -0.490
Weights for window positions 1,..,9, using linear weight variation model:
1 2 3 4 5 6 7 8 9
1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00
edge center edge
Fig 1: Hydropathy plot of Glucuronidase protein
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

9PROTEIN DISCOVERY AND ANALYSIS
Source: ProtScale
A hydropathy plot allows the visualisation of hydrophobicity over the complete peptide
length sequence. The hydropathy scale has been found to be based on the hydrophilic and
hydrophobic properties of all the 20 amino acids which have been found to be the main
composition of the protein used in this report (Johri et al., 2019). The highest peak value has
been observed as a positive peak in 450 which states that the amino acid is well inside the
protein core. Thus, it can be stated that this amino acid is inside the hydrophobic core of the
protein. The highest negative peak has been observed at 30 position which proves that the
amino acid located at this region is well situated outside the protein. This amino acid has
been found to be located near the hydrophilic surroundings of the protein which states that it
may be near the surround water. This is the overall details which can be drawn out from the
hydropathy plot given above.
Initial purification of the protein
Initial purification of beta glucuronidase was done from human placenta (Brot et al., 1978).
The purification value was 18,000 fold homogenous solution which was divided in three
steps-
1. Immunoadsorption- This process was performed on antibody-Sepharose resin.
2. DEAE-Sephadex chromatography- This process was done to separate the protein from the
total composition
3. PAGE (Poly acrylamide gel electrophoresis) was performed.
4. Sedimentation against rat and goat antibody was checked.
5. Finally, it was confirmed that the protein was beta glucuronidase
Source: ProtScale
A hydropathy plot allows the visualisation of hydrophobicity over the complete peptide
length sequence. The hydropathy scale has been found to be based on the hydrophilic and
hydrophobic properties of all the 20 amino acids which have been found to be the main
composition of the protein used in this report (Johri et al., 2019). The highest peak value has
been observed as a positive peak in 450 which states that the amino acid is well inside the
protein core. Thus, it can be stated that this amino acid is inside the hydrophobic core of the
protein. The highest negative peak has been observed at 30 position which proves that the
amino acid located at this region is well situated outside the protein. This amino acid has
been found to be located near the hydrophilic surroundings of the protein which states that it
may be near the surround water. This is the overall details which can be drawn out from the
hydropathy plot given above.
Initial purification of the protein
Initial purification of beta glucuronidase was done from human placenta (Brot et al., 1978).
The purification value was 18,000 fold homogenous solution which was divided in three
steps-
1. Immunoadsorption- This process was performed on antibody-Sepharose resin.
2. DEAE-Sephadex chromatography- This process was done to separate the protein from the
total composition
3. PAGE (Poly acrylamide gel electrophoresis) was performed.
4. Sedimentation against rat and goat antibody was checked.
5. Finally, it was confirmed that the protein was beta glucuronidase
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
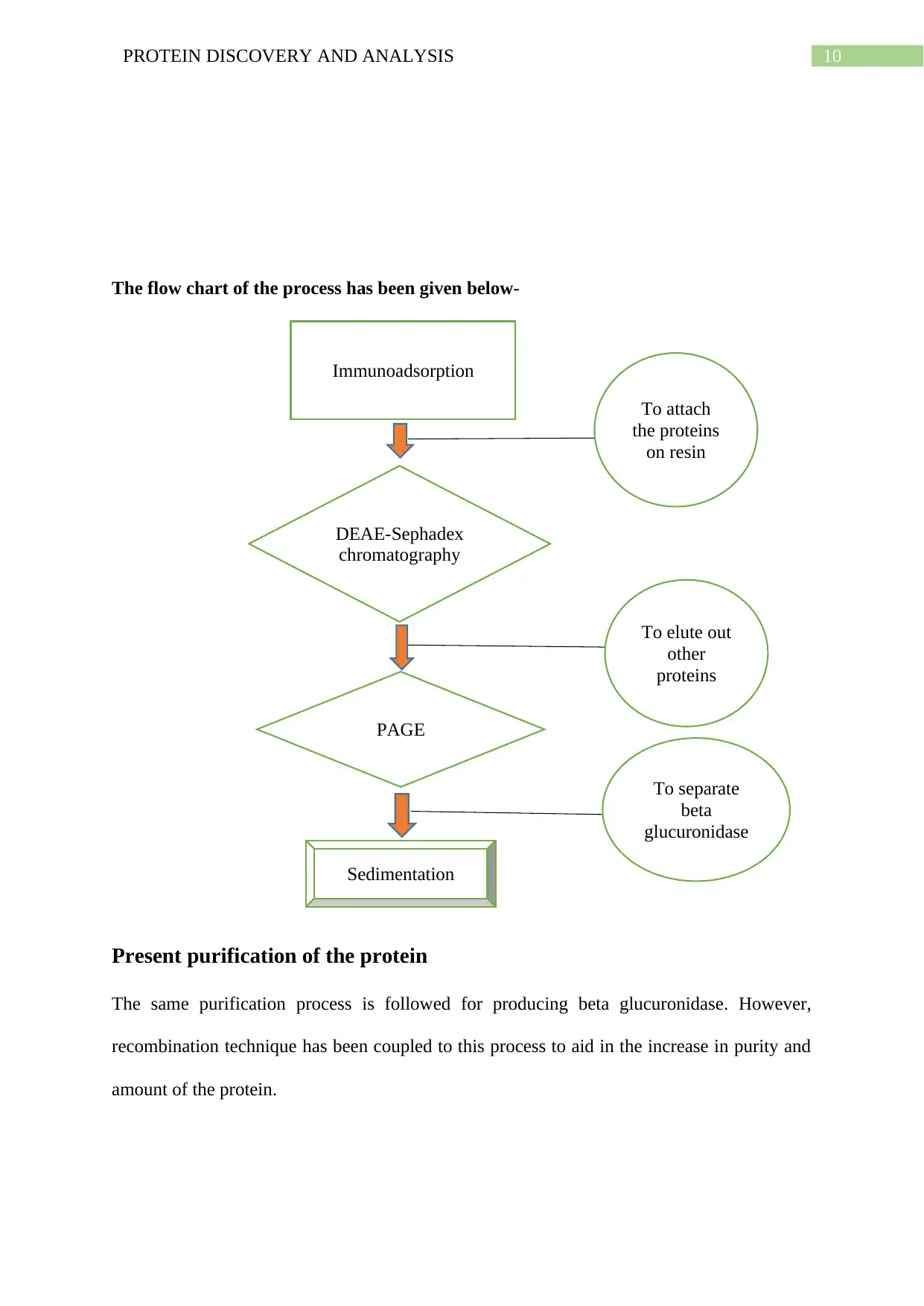
10PROTEIN DISCOVERY AND ANALYSIS
The flow chart of the process has been given below-
Present purification of the protein
The same purification process is followed for producing beta glucuronidase. However,
recombination technique has been coupled to this process to aid in the increase in purity and
amount of the protein.
Immunoadsorption
DEAE-Sephadex
chromatography
PAGE
Sedimentation
To attach
the proteins
on resin
To elute out
other
proteins
To separate
beta
glucuronidase
The flow chart of the process has been given below-
Present purification of the protein
The same purification process is followed for producing beta glucuronidase. However,
recombination technique has been coupled to this process to aid in the increase in purity and
amount of the protein.
Immunoadsorption
DEAE-Sephadex
chromatography
PAGE
Sedimentation
To attach
the proteins
on resin
To elute out
other
proteins
To separate
beta
glucuronidase

11PROTEIN DISCOVERY AND ANALYSIS
Flow chart of the process:
Isocitarte
dehydrogenase
promoter is added to
the E. coli genome.
Enzyme
extraction
Ammonium
sulfate
precipitation
DEAE-Ion
exchange
chromatography
Hydroxyapatite steric ion-
exchange chromatography.
Protein
separation
Isolation of
the protein
from mixture
Flow chart of the process:
Isocitarte
dehydrogenase
promoter is added to
the E. coli genome.
Enzyme
extraction
Ammonium
sulfate
precipitation
DEAE-Ion
exchange
chromatography
Hydroxyapatite steric ion-
exchange chromatography.
Protein
separation
Isolation of
the protein
from mixture
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 14
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.
