Extraction of Disease Surveillance Using Big Data in CTIS Research S1
VerifiedAdded on 2023/03/31
|6
|881
|161
Report
AI Summary
This report discusses the extraction of disease surveillance using big data analysis, focusing on influenza disease prediction. The method involves collecting data from social media and internet searches to analyze health-related statistics. The analysis, performed using SPSS, shows strong correlations between social media data and surveillance data from health organizations. The system effectively detects weekly changes in influenza prevalence, demonstrating the utility of distinguishing infection-related tweets. A regression analysis model is applied to predict disease stroke visits based on near-real-time environmental and social media data, with approximately 80% precision, aiding in preventative measures and treatment.

Running head: EXTRACTION OF DISEASE SURVEILLANCE USING BIG DATA 1
Extraction of Disease Surveillance Using Big Data
By (Name of Student)
(Institutional Affiliation)
(Date of Submission)
(Course Code)
Extraction of Disease Surveillance Using Big Data
By (Name of Student)
(Institutional Affiliation)
(Date of Submission)
(Course Code)
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

EXTRACTION OF DISEASE SURVEILLANCE USING BIG DATA 2
Extraction of Disease Surveillance Using Big Data
Research Progress/ Method of Investigation
Big data can be used for analysis for extraction of disease surveillance. Today, many
users of social networking shares their health related statistics on web. Such health related
information can use for extraction of diseases (Thompson et al., 2010). Diseases such as
blood pressure, diabetes are most rampant and costly chronic conditions in the world which
cannot be cured. However accurate and timely surveillance data can control these diseases
(Martin-Sanchez, 2014). The aim of this work is to provide results of the data analysis based
on the surveillance of influenza disease using big data. In this paper, therefore, generalized
method is discussed which will extract the fore-coming strokes of different diseases
depending on data. Here data is collected from social media, internet search. This method was
opted since it is a popular data source for disease surveillance using social media since it can
provide nearly instant access to real-time social opinions.
Results/Findings
The objective was to analyze the data collected from social media and internet sources such
as twitter and Facebook to predict surveillance of influenza disease during the most recent
2016–2019 flu season and to analyze the performance at multiple levels of geographic
granularity, unlike past studies that focused on general or regional surveillance. The analysis
of the data was done using Statistical Packages for Social Sciences (SPSS Version 25.0).
Based on the analysis, the estimates of influenza prevalence were strongly correlated with
surveillance data from the Centres for Disease Control and Anticipation for the Health
ministry (r = 0.83, p < 0.001) as well as investigation data from the Department of Health and
Mental Hygiene of (r = 0.92, p < 0.001). Our system detected the weekly change in direction
Extraction of Disease Surveillance Using Big Data
Research Progress/ Method of Investigation
Big data can be used for analysis for extraction of disease surveillance. Today, many
users of social networking shares their health related statistics on web. Such health related
information can use for extraction of diseases (Thompson et al., 2010). Diseases such as
blood pressure, diabetes are most rampant and costly chronic conditions in the world which
cannot be cured. However accurate and timely surveillance data can control these diseases
(Martin-Sanchez, 2014). The aim of this work is to provide results of the data analysis based
on the surveillance of influenza disease using big data. In this paper, therefore, generalized
method is discussed which will extract the fore-coming strokes of different diseases
depending on data. Here data is collected from social media, internet search. This method was
opted since it is a popular data source for disease surveillance using social media since it can
provide nearly instant access to real-time social opinions.
Results/Findings
The objective was to analyze the data collected from social media and internet sources such
as twitter and Facebook to predict surveillance of influenza disease during the most recent
2016–2019 flu season and to analyze the performance at multiple levels of geographic
granularity, unlike past studies that focused on general or regional surveillance. The analysis
of the data was done using Statistical Packages for Social Sciences (SPSS Version 25.0).
Based on the analysis, the estimates of influenza prevalence were strongly correlated with
surveillance data from the Centres for Disease Control and Anticipation for the Health
ministry (r = 0.83, p < 0.001) as well as investigation data from the Department of Health and
Mental Hygiene of (r = 0.92, p < 0.001). Our system detected the weekly change in direction
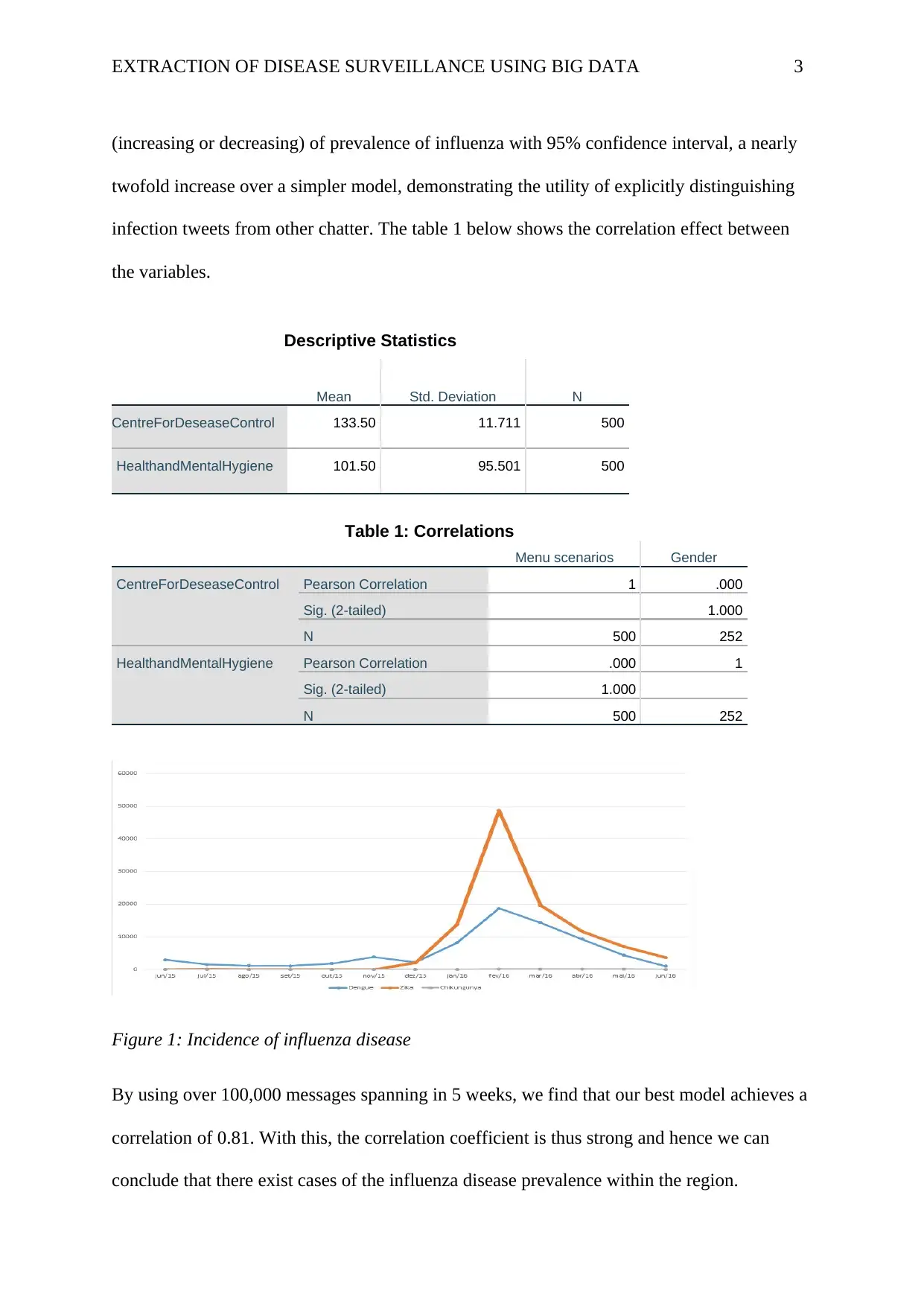
EXTRACTION OF DISEASE SURVEILLANCE USING BIG DATA 3
(increasing or decreasing) of prevalence of influenza with 95% confidence interval, a nearly
twofold increase over a simpler model, demonstrating the utility of explicitly distinguishing
infection tweets from other chatter. The table 1 below shows the correlation effect between
the variables.
Descriptive Statistics
Mean Std. Deviation N
CentreForDeseaseControl 133.50 11.711 500
HealthandMentalHygiene 101.50 95.501 500
Table 1: Correlations
Menu scenarios Gender
CentreForDeseaseControl Pearson Correlation 1 .000
Sig. (2-tailed) 1.000
N 500 252
HealthandMentalHygiene Pearson Correlation .000 1
Sig. (2-tailed) 1.000
N 500 252
Figure 1: Incidence of influenza disease
By using over 100,000 messages spanning in 5 weeks, we find that our best model achieves a
correlation of 0.81. With this, the correlation coefficient is thus strong and hence we can
conclude that there exist cases of the influenza disease prevalence within the region.
(increasing or decreasing) of prevalence of influenza with 95% confidence interval, a nearly
twofold increase over a simpler model, demonstrating the utility of explicitly distinguishing
infection tweets from other chatter. The table 1 below shows the correlation effect between
the variables.
Descriptive Statistics
Mean Std. Deviation N
CentreForDeseaseControl 133.50 11.711 500
HealthandMentalHygiene 101.50 95.501 500
Table 1: Correlations
Menu scenarios Gender
CentreForDeseaseControl Pearson Correlation 1 .000
Sig. (2-tailed) 1.000
N 500 252
HealthandMentalHygiene Pearson Correlation .000 1
Sig. (2-tailed) 1.000
N 500 252
Figure 1: Incidence of influenza disease
By using over 100,000 messages spanning in 5 weeks, we find that our best model achieves a
correlation of 0.81. With this, the correlation coefficient is thus strong and hence we can
conclude that there exist cases of the influenza disease prevalence within the region.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
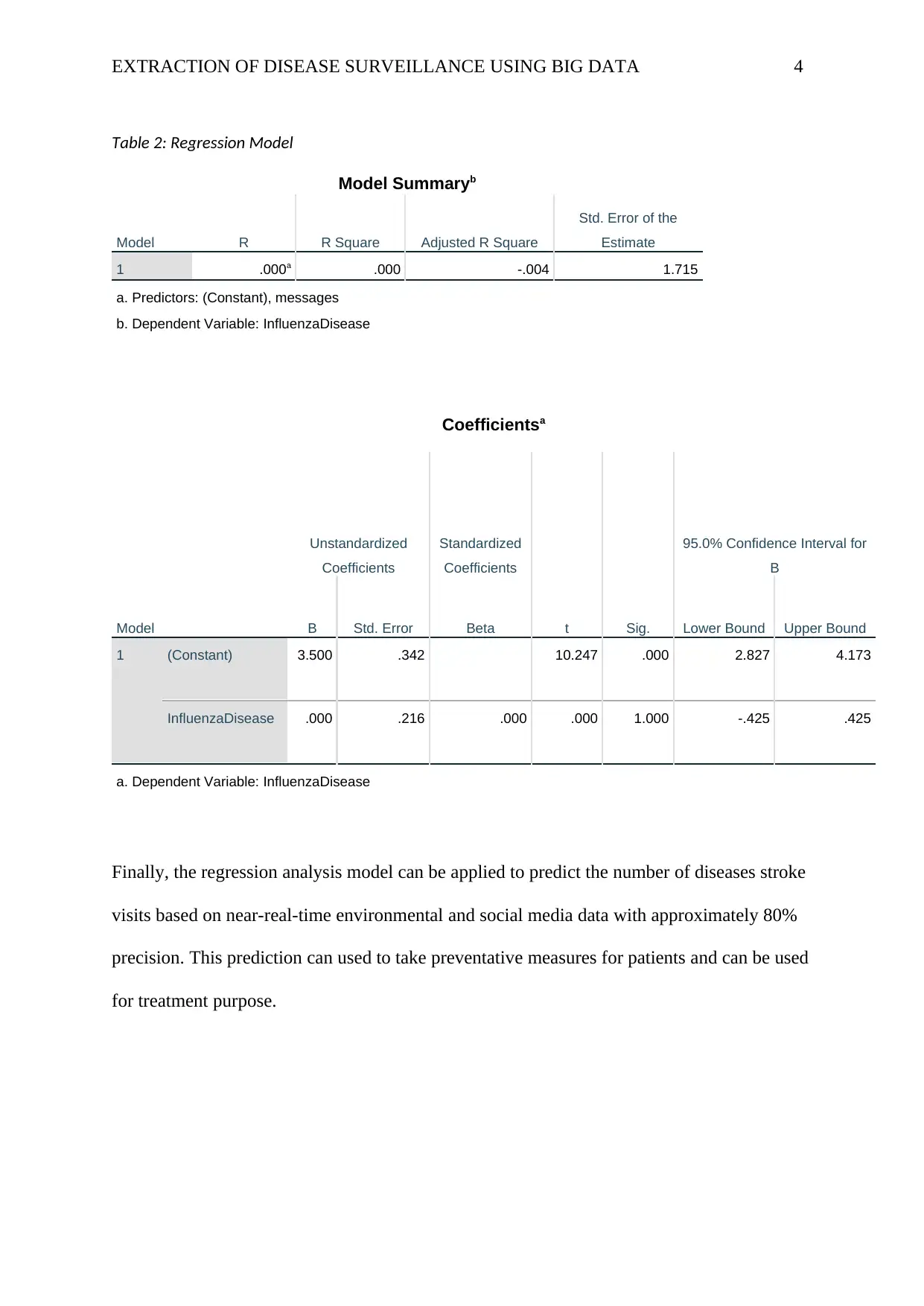
EXTRACTION OF DISEASE SURVEILLANCE USING BIG DATA 4
Table 2: Regression Model
Model Summaryb
Model R R Square Adjusted R Square
Std. Error of the
Estimate
1 .000a .000 -.004 1.715
a. Predictors: (Constant), messages
b. Dependent Variable: InfluenzaDisease
Coefficientsa
Model
Unstandardized
Coefficients
Standardized
Coefficients
t Sig.
95.0% Confidence Interval for
B
B Std. Error Beta Lower Bound Upper Bound
1 (Constant) 3.500 .342 10.247 .000 2.827 4.173
InfluenzaDisease .000 .216 .000 .000 1.000 -.425 .425
a. Dependent Variable: InfluenzaDisease
Finally, the regression analysis model can be applied to predict the number of diseases stroke
visits based on near-real-time environmental and social media data with approximately 80%
precision. This prediction can used to take preventative measures for patients and can be used
for treatment purpose.
Table 2: Regression Model
Model Summaryb
Model R R Square Adjusted R Square
Std. Error of the
Estimate
1 .000a .000 -.004 1.715
a. Predictors: (Constant), messages
b. Dependent Variable: InfluenzaDisease
Coefficientsa
Model
Unstandardized
Coefficients
Standardized
Coefficients
t Sig.
95.0% Confidence Interval for
B
B Std. Error Beta Lower Bound Upper Bound
1 (Constant) 3.500 .342 10.247 .000 2.827 4.173
InfluenzaDisease .000 .216 .000 .000 1.000 -.425 .425
a. Dependent Variable: InfluenzaDisease
Finally, the regression analysis model can be applied to predict the number of diseases stroke
visits based on near-real-time environmental and social media data with approximately 80%
precision. This prediction can used to take preventative measures for patients and can be used
for treatment purpose.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

EXTRACTION OF DISEASE SURVEILLANCE USING BIG DATA 5
References
Eick, A. A., Uyeki, T. M., Klimov, A., Hall, H., Reid, R., Santosham, M., & O’brien, K. L.
(2011). Maternal influenza vaccination and effect on influenza virus infection in
young infants. Archives of pediatrics & adolescent medicine, 165(2), 104-111.
Hayward, A. C., Fragaszy, E. B., Bermingham, A., Wang, L., Copas, A., Edmunds, W. J., ...
& Lim, M. S. (2014). Comparative community burden and severity of seasonal and
pandemic influenza: results of the Flu Watch cohort study. The Lancet Respiratory
Medicine, 2(6), 445-454.
Larwood, L. (1978). Swine flu: A field study of self‐serving biases. Journal of Applied Social
Psychology, 8(3), 283-289.
Lynberg, M. C., Khoury, M. J., Lu, X., & Cocian, T. (1994). Maternal flu, fever, and the risk
of neural tube defects: a population-based case-control study. American journal of
epidemiology, 140(3), 244-255.
Manogaran, G., & Lopez, D. (2018). Disease surveillance system for big climate data
processing and dengue transmission. In Climate Change and Environmental
Concerns: Breakthroughs in Research and Practice (pp. 427-446). IGI Global.
Martin-Sanchez, F., & Verspoor, K. (2014). Big data in medicine is driving big
changes. Yearbook of medical informatics, 23(01), 14-20.
Ram, S., Zhang, W., Williams, M., & Pengetnze, Y. (2015). Predicting asthma-related
emergency department visits using big data. IEEE journal of biomedical and health
informatics, 19(4), 1216-1223.
References
Eick, A. A., Uyeki, T. M., Klimov, A., Hall, H., Reid, R., Santosham, M., & O’brien, K. L.
(2011). Maternal influenza vaccination and effect on influenza virus infection in
young infants. Archives of pediatrics & adolescent medicine, 165(2), 104-111.
Hayward, A. C., Fragaszy, E. B., Bermingham, A., Wang, L., Copas, A., Edmunds, W. J., ...
& Lim, M. S. (2014). Comparative community burden and severity of seasonal and
pandemic influenza: results of the Flu Watch cohort study. The Lancet Respiratory
Medicine, 2(6), 445-454.
Larwood, L. (1978). Swine flu: A field study of self‐serving biases. Journal of Applied Social
Psychology, 8(3), 283-289.
Lynberg, M. C., Khoury, M. J., Lu, X., & Cocian, T. (1994). Maternal flu, fever, and the risk
of neural tube defects: a population-based case-control study. American journal of
epidemiology, 140(3), 244-255.
Manogaran, G., & Lopez, D. (2018). Disease surveillance system for big climate data
processing and dengue transmission. In Climate Change and Environmental
Concerns: Breakthroughs in Research and Practice (pp. 427-446). IGI Global.
Martin-Sanchez, F., & Verspoor, K. (2014). Big data in medicine is driving big
changes. Yearbook of medical informatics, 23(01), 14-20.
Ram, S., Zhang, W., Williams, M., & Pengetnze, Y. (2015). Predicting asthma-related
emergency department visits using big data. IEEE journal of biomedical and health
informatics, 19(4), 1216-1223.

EXTRACTION OF DISEASE SURVEILLANCE USING BIG DATA 6
Viboud, C., Alonso, W. J., & Simonsen, L. (2006). Influenza in tropical regions. PLoS
medicine, 3(4), e89.
Thompson, M. G., Shay, D. K., Zhou, H., Bridges, C. B., Cheng, P. Y., Burns, E., ... & Cox,
N. J. (2010). Estimates of deaths associated with seasonal influenza-United States,
1976-2007. Morbidity and Mortality Weekly Report, 59(33), 1057-1062.
Viboud, C., Alonso, W. J., & Simonsen, L. (2006). Influenza in tropical regions. PLoS
medicine, 3(4), e89.
Thompson, M. G., Shay, D. K., Zhou, H., Bridges, C. B., Cheng, P. Y., Burns, E., ... & Cox,
N. J. (2010). Estimates of deaths associated with seasonal influenza-United States,
1976-2007. Morbidity and Mortality Weekly Report, 59(33), 1057-1062.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 6
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.




