Exploring Competition Dialysis for Ligand Discovery in Biochemistry
VerifiedAdded on 2023/06/04
|5
|714
|478
Homework Assignment
AI Summary
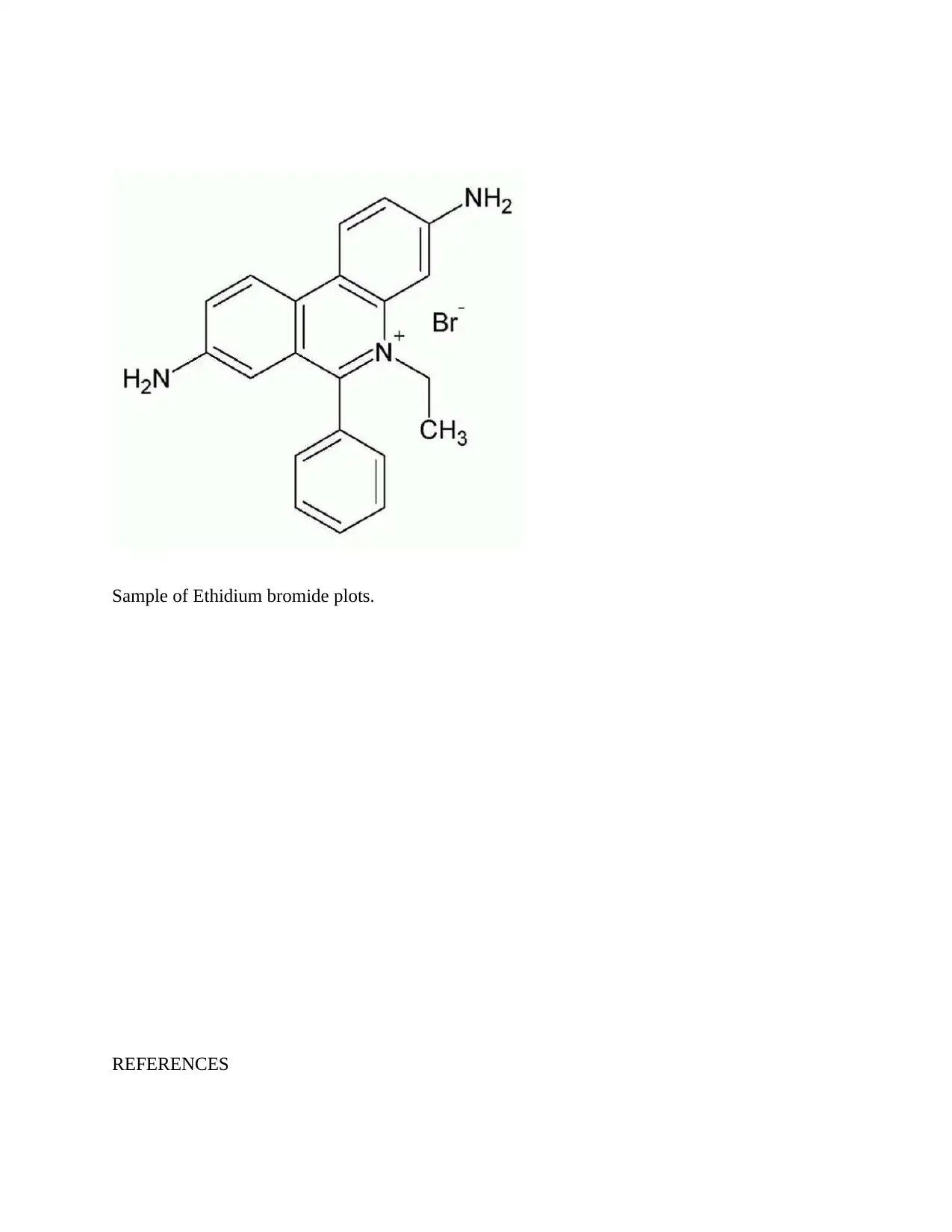
This assignment focuses on competition dialysis, a technique used in biochemistry for discovering ligands that bind to nucleic acids with sequence selectivity. It discusses Brad Chaires' method for competition dialysis, including solution preparation, concentrations used (specifically 1uM and 0.75uM), and the purpose of adding surfactants to improve wetting properties. The assignment identifies fluorescence spectroscopy as the method used for determining ligand concentration due to its parameter adjustability. Data analysis examples include studies with quadruplex forms and ligands like methylene blue and ethidium, detailing binding preferences and energy calculations. The document concludes with examples of selectivity plots, particularly for ethidium bromide, and provides relevant references.
1 out of 5






![[object Object]](/_next/static/media/star-bottom.7253800d.svg)