BIOINFORMATICS ASSIGNMENT 1: Analysis of Genes and Protein Sequences
VerifiedAdded on 2021/06/18
|56
|10564
|54
Homework Assignment
AI Summary
This bioinformatics assignment encompasses a comprehensive exploration of fundamental concepts and practical applications within the field. The assignment begins with an introduction to key bioinformatics terms, including BLAST, GenBank, Ensembl, GEO, Pfam, KEGG, OMIM, PDB, GO, and R, establishing a foundational understanding of the tools and databases used in biological sequence analysis. The core of the assignment involves DNA sequence translation, including determining the correct open reading frame (ORF), explaining the presence of multiple reading frames, identifying encoded proteins, and understanding protein families and functions. The assignment also delves into sequence homology, defining terms like homologue, orthologue, and paralogue, and exploring concepts of score, E-value, identity, and conservative substitution within protein alignments. Furthermore, the assignment emphasizes practical skills by guiding students through the retrieval of gene, mRNA, and protein information from the NCBI database, using the BRCA2 gene as a case study. The assignment requires the student to acquire and analyze sequence information from various databases, perform basic analysis of evolutionarily conserved sequences, and conduct multiple sequence alignments across different species. Finally, the assignment concludes with the use of the PDB database for THREE-DIMENSIONAL VIEWING OF AN IDENTIFIED PROTEIN STRUCTURE.

BIOINFORMATICS ASSIGNMENT 1
I
declare that all material in this assessment is my own work except where there is clear
acknowledgement and reference to the work of others. I have read the Academic Honesty and Assessment
Obligations for Coursework Students Policy and Academic Dishonesty Procedures
(http://www.adelaide.edu.au/policies/230/).
I give permission for my assessment work to be reproduced and submitted to other academic staff
for the purposes of assessment and to be copied, submitted and retained in a form suitable for
electronic checking of plagiarism.
Signed………………………………………………. Date ……………………………………………
Table of Contents
A. Introductory Bioinformatics........................................................................................................................2
1. Terms commonly used in bioinformatics....................................................................................................2
2. DNA Sequence Translation.........................................................................................................................4
3. Sequence homology.....................................................................................................................................6
4. Learn To Retrieve Gene/mRNA/Protein Information.................................................................................9
B. Genetic Analysis of a Human Cancer Disease Using Databases.................................................................35
School of Biological Sciences
Assessment Cover Sheet
Student Name
Student ID
Assessment Title
Course/Program
Lecturer/Tutor
Date Submitted
OFFICE USE ONLY
Date Received
I
declare that all material in this assessment is my own work except where there is clear
acknowledgement and reference to the work of others. I have read the Academic Honesty and Assessment
Obligations for Coursework Students Policy and Academic Dishonesty Procedures
(http://www.adelaide.edu.au/policies/230/).
I give permission for my assessment work to be reproduced and submitted to other academic staff
for the purposes of assessment and to be copied, submitted and retained in a form suitable for
electronic checking of plagiarism.
Signed………………………………………………. Date ……………………………………………
Table of Contents
A. Introductory Bioinformatics........................................................................................................................2
1. Terms commonly used in bioinformatics....................................................................................................2
2. DNA Sequence Translation.........................................................................................................................4
3. Sequence homology.....................................................................................................................................6
4. Learn To Retrieve Gene/mRNA/Protein Information.................................................................................9
B. Genetic Analysis of a Human Cancer Disease Using Databases.................................................................35
School of Biological Sciences
Assessment Cover Sheet
Student Name
Student ID
Assessment Title
Course/Program
Lecturer/Tutor
Date Submitted
OFFICE USE ONLY
Date Received
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

BIOINFORMATICS ASSIGNMENT 2
5. Analysis of a Human Genetic Disease......................................................................................................35
6. Acquiring Sequence Information.............................................................................................................37
7. Uniprot/Swiss-Prot Database..................................................................................................................42
8. Basic Analysis of Evolutionarily Conserved Sequences........................................................................43
9. Multiple Sequence Alignments across Different Species......................................................................45
10. THREE-DIMENSIONAL VIEWING OF AN IDENTIFIED PROTEIN STRUCTURE:................50
References.........................................................................................................................................................54
5. Analysis of a Human Genetic Disease......................................................................................................35
6. Acquiring Sequence Information.............................................................................................................37
7. Uniprot/Swiss-Prot Database..................................................................................................................42
8. Basic Analysis of Evolutionarily Conserved Sequences........................................................................43
9. Multiple Sequence Alignments across Different Species......................................................................45
10. THREE-DIMENSIONAL VIEWING OF AN IDENTIFIED PROTEIN STRUCTURE:................50
References.........................................................................................................................................................54

BIOINFORMATICS ASSIGNMENT 3
A. Introductory Bioinformatics
1. Terms commonly used in bioinformatics
BLAST
Basic Local Alignment Search Tool (BLAST) finds regions of local similarity among
sequences. It compares nucleotide or protein sequences to sequence database and calculate the
statistical significance of matched results.
GenBank
It is a genetic sequence database. GenBank stores annotated collection of DNA sequences
which is publically available. It is a part of the International nucleotide sequence database
collaboration. It is designed to assess and encourage access within the scientific community to the
updated and comprehensive DNA sequence information.
Ensembl
Ensembl is a genome browser for the vertebrate genomes that used to support research in
comparative genomes, evaluation, sequence variation and transcriptional regulation. Ensembl annotate
gene, it computes multiple alignment, predicts regulatory function and stores data of various diseases.
Ensembl includes tools like BLAST, BLAT, BioMart and a variant effect predictor for all supported
species.
GEO
Gene Expression Omnibus (GEO) is an international public repository which archives and
freely distributes microarray, next generation sequencing and other forms of high throughout functional
A. Introductory Bioinformatics
1. Terms commonly used in bioinformatics
BLAST
Basic Local Alignment Search Tool (BLAST) finds regions of local similarity among
sequences. It compares nucleotide or protein sequences to sequence database and calculate the
statistical significance of matched results.
GenBank
It is a genetic sequence database. GenBank stores annotated collection of DNA sequences
which is publically available. It is a part of the International nucleotide sequence database
collaboration. It is designed to assess and encourage access within the scientific community to the
updated and comprehensive DNA sequence information.
Ensembl
Ensembl is a genome browser for the vertebrate genomes that used to support research in
comparative genomes, evaluation, sequence variation and transcriptional regulation. Ensembl annotate
gene, it computes multiple alignment, predicts regulatory function and stores data of various diseases.
Ensembl includes tools like BLAST, BLAT, BioMart and a variant effect predictor for all supported
species.
GEO
Gene Expression Omnibus (GEO) is an international public repository which archives and
freely distributes microarray, next generation sequencing and other forms of high throughout functional
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

BIOINFORMATICS ASSIGNMENT 4
genomic data which is submitted by the research community tools. These tools are provided to help
users query and download experiments and curated gene expression profile.
Pfam
Pfam is the database includes large collections of protein families, represented by multiple
sequence alignments and HMMs (Hidden Markov Model). It is available on World Wide Web and
provides higher level groupings of the related entries, called clans which are the collection of Pfam
entries.
KEGG
Kyoto Encyclopedia of Genes and Genomes (KEGG) is the collection of databases deals with
genomes, biological pathways, disease drugs and the chemical substances. KEGG is a database
resource to understanding the high level functions. It utilities the biological system, such as cell, the
organism and ecosystem from molecular- level information, specifically large scale molecular database
generated by genome sequencing and other high throughout experimental technologies.
OMIM
Online Mendelian Inheritance in Man (OMIM) is the database initiated by Dr. Victor A.
Mckusick in 1960. It is a comprehensive, authoritative compendium of human’s genes and genome
which is freely available and updated routinely.
PDB
Protein Data Bank or PDB is the open access digital data resources in all biology and medicine.
It is in demand for experimental data central to the scientific discovery. PDB provides access to the 3D
structure data for large molecules like protein, DNA and RNA to understand its role in human and
animal health.
genomic data which is submitted by the research community tools. These tools are provided to help
users query and download experiments and curated gene expression profile.
Pfam
Pfam is the database includes large collections of protein families, represented by multiple
sequence alignments and HMMs (Hidden Markov Model). It is available on World Wide Web and
provides higher level groupings of the related entries, called clans which are the collection of Pfam
entries.
KEGG
Kyoto Encyclopedia of Genes and Genomes (KEGG) is the collection of databases deals with
genomes, biological pathways, disease drugs and the chemical substances. KEGG is a database
resource to understanding the high level functions. It utilities the biological system, such as cell, the
organism and ecosystem from molecular- level information, specifically large scale molecular database
generated by genome sequencing and other high throughout experimental technologies.
OMIM
Online Mendelian Inheritance in Man (OMIM) is the database initiated by Dr. Victor A.
Mckusick in 1960. It is a comprehensive, authoritative compendium of human’s genes and genome
which is freely available and updated routinely.
PDB
Protein Data Bank or PDB is the open access digital data resources in all biology and medicine.
It is in demand for experimental data central to the scientific discovery. PDB provides access to the 3D
structure data for large molecules like protein, DNA and RNA to understand its role in human and
animal health.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

BIOINFORMATICS ASSIGNMENT 5
GO
Gene Ontology (GO) is the major bioinformatics initiative to create a computational
representation of peoples evolving knowledge to understand how genes encode function at the
molecular, cellular and tissue system levels. GO resources plays an important role to support
biomedical research, which includes interpretation of the large scale molecular experiments and
computational analysis of biological knowledge.
R
R is the comprehensive statistical environment and a programming language for professional
data analysis and graphical display. Bio-conductor project which associated with R provides various
additional R packages for statistical data analysis in different fields of life science, such as tools for
microarray, next generation sequence and genome analysis. R software is freely available and works in
all common operating systems.
2. DNA Sequence Translation
A.
What is the correct open reading frame?
MADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNM
PNMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKL
NKIFEKLGM
Why are there 6 reading frames instead of 3?
The coding strand refers to a DNA strand with the same base order like the RNA transcript for a
particular gene. Because one gene is always entirely present on a single DNA strand there are indeed 3
reading frames possibly present in this strand, and only 1 RF actually containing the correct codon
sequence for this gene, however when the entire genome is viewed, it is possible for one gene to be
GO
Gene Ontology (GO) is the major bioinformatics initiative to create a computational
representation of peoples evolving knowledge to understand how genes encode function at the
molecular, cellular and tissue system levels. GO resources plays an important role to support
biomedical research, which includes interpretation of the large scale molecular experiments and
computational analysis of biological knowledge.
R
R is the comprehensive statistical environment and a programming language for professional
data analysis and graphical display. Bio-conductor project which associated with R provides various
additional R packages for statistical data analysis in different fields of life science, such as tools for
microarray, next generation sequence and genome analysis. R software is freely available and works in
all common operating systems.
2. DNA Sequence Translation
A.
What is the correct open reading frame?
MADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNM
PNMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKL
NKIFEKLGM
Why are there 6 reading frames instead of 3?
The coding strand refers to a DNA strand with the same base order like the RNA transcript for a
particular gene. Because one gene is always entirely present on a single DNA strand there are indeed 3
reading frames possibly present in this strand, and only 1 RF actually containing the correct codon
sequence for this gene, however when the entire genome is viewed, it is possible for one gene to be

BIOINFORMATICS ASSIGNMENT 6
available on single strand with another gene present on other which means that the coding strand for
single gene is the non-coding strand for the other. That is why there are 6 reading frames are found in
result for the genome as a whole as both strands contain genes.
Save and paste the protein sequence you obtain from the “Compact M- no space” format here?
Answer:-
5'3' Frame 1
MADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNM
PNMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKL
NKIFEKLGM-
5'3' Frame 2
WRIKNLNFWLWMTFPPCDA-CVTC-KSWDSIMLRKRKMASTLSISCRQAVMDLLSPTGTC
PIWMAWNC-KQFVRMARCRHCQC-W-LQKRRKRTSLLRRKRGPVAMW-SHLPPRRWRKNS
TKSLRNWAC
5'3' Frame 3
GG-RT-IFGCG-LFHHATHSA-PAERAGIQ-C-GSGRWRRRSQ-VAGRRLWICYLRLEHA
QYGWPGIAENNSCGWRDVGIASVNGDCRSEEREHHCCGASGGQWLCGEAIYRRDAGGKTQ
QNL-ETGHV
3'5' Frame 1
SHAQFLKDFVEFFLQRRGGKWLHHIATGPRLRRSNDVLFLRFCSHH-HWQCRHRAIRTNC
FQQFQAIHIGHVPVGDNKSITACLQLIESVDAIFRFLNIIESQLFQQVTHYASHGGKVIH
NQKFKFFIRH
3'5' Frame 2
HMPSFSKILLSFSSSVAAVNGFTT-PLAPACAAAMMFSFFASAVTINTGNADIAPSARIV
FSNSRPSILGMFQSEITNP-PPACNLLRASTPSSASSTLLNPSSFSRLRTMRRMVEKSST
TKNLSSLSA
3'5' Frame 3
TCPVSQRFC-VFPPASRR-MASPHSHWPPLAPQQ-CSLSSLLQSPLTLAMPTSRHPHELF
SAIPGHPYWACSSRR-QIHNRLPATY-ERRRHLPLPQHY-IPALSAGYALCVAWWKSHPQ
PKI-VLYPP
B
What is the protein encoded by the ORF you identified and what species is it from?
Answer- Chemotaxis protein
Species- Escherichia Coli
available on single strand with another gene present on other which means that the coding strand for
single gene is the non-coding strand for the other. That is why there are 6 reading frames are found in
result for the genome as a whole as both strands contain genes.
Save and paste the protein sequence you obtain from the “Compact M- no space” format here?
Answer:-
5'3' Frame 1
MADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNM
PNMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKL
NKIFEKLGM-
5'3' Frame 2
WRIKNLNFWLWMTFPPCDA-CVTC-KSWDSIMLRKRKMASTLSISCRQAVMDLLSPTGTC
PIWMAWNC-KQFVRMARCRHCQC-W-LQKRRKRTSLLRRKRGPVAMW-SHLPPRRWRKNS
TKSLRNWAC
5'3' Frame 3
GG-RT-IFGCG-LFHHATHSA-PAERAGIQ-C-GSGRWRRRSQ-VAGRRLWICYLRLEHA
QYGWPGIAENNSCGWRDVGIASVNGDCRSEEREHHCCGASGGQWLCGEAIYRRDAGGKTQ
QNL-ETGHV
3'5' Frame 1
SHAQFLKDFVEFFLQRRGGKWLHHIATGPRLRRSNDVLFLRFCSHH-HWQCRHRAIRTNC
FQQFQAIHIGHVPVGDNKSITACLQLIESVDAIFRFLNIIESQLFQQVTHYASHGGKVIH
NQKFKFFIRH
3'5' Frame 2
HMPSFSKILLSFSSSVAAVNGFTT-PLAPACAAAMMFSFFASAVTINTGNADIAPSARIV
FSNSRPSILGMFQSEITNP-PPACNLLRASTPSSASSTLLNPSSFSRLRTMRRMVEKSST
TKNLSSLSA
3'5' Frame 3
TCPVSQRFC-VFPPASRR-MASPHSHWPPLAPQQ-CSLSSLLQSPLTLAMPTSRHPHELF
SAIPGHPYWACSSRR-QIHNRLPATY-ERRRHLPLPQHY-IPALSAGYALCVAWWKSHPQ
PKI-VLYPP
B
What is the protein encoded by the ORF you identified and what species is it from?
Answer- Chemotaxis protein
Species- Escherichia Coli
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

BIOINFORMATICS ASSIGNMENT 7
To what family of protein does it belongs?
Answer- Family- CheY
What is the function of this protein?
Answer- It Involved in transmit the sensory signals from chemoreceptors of the flagellar motors. This
protein is also participated in changing the direction of flagellar rotation.
3. Sequence homology
Define the terms
Homologue: - A chromosome which is similar in physical aspects and genetic information to another
chromosome, where both make pairs during meiosis.
Orthologue: - Sequence that have common ancestor and have divided due to speciation event
Paralogue: - Sequences are the descendants of an ancestral gene that underwent a duplication event.
List the details of the each of these homologous sequences here, including its maximum identity
Answer- Homologous sequence
1 adkelkflvv ddestmrriv rnllkelgfn nveeaedgvd alnklqaggy gfvisdwmmp
61 nmdglellkt iradgamsal pvlmvtalak keniiaaaqa gasgyvvkpf taatleekln
121 kifeklgm
Accession no. 3F7N_A
SOURCE ESCHERIA COLI K-12
AMINO ACID- 128
MAX SCORE – 246
MAX IDENT 98%
1 adkelkflvv ddfstmrriv rnllkelgfn nveeaedgvd alnklqaggf gfiicdwnmp
61 nmdglellkt iradsamsal pvlmvtaeak keniiaaaqa gasgyvvkpf taatleekln
121 kifeklgm
Accession no.- 2CHY_A
Source- Salmonella Enterica Subsp. Eterica Serovar Typhimurium
Amino Acid- 128
Max score- 251
To what family of protein does it belongs?
Answer- Family- CheY
What is the function of this protein?
Answer- It Involved in transmit the sensory signals from chemoreceptors of the flagellar motors. This
protein is also participated in changing the direction of flagellar rotation.
3. Sequence homology
Define the terms
Homologue: - A chromosome which is similar in physical aspects and genetic information to another
chromosome, where both make pairs during meiosis.
Orthologue: - Sequence that have common ancestor and have divided due to speciation event
Paralogue: - Sequences are the descendants of an ancestral gene that underwent a duplication event.
List the details of the each of these homologous sequences here, including its maximum identity
Answer- Homologous sequence
1 adkelkflvv ddestmrriv rnllkelgfn nveeaedgvd alnklqaggy gfvisdwmmp
61 nmdglellkt iradgamsal pvlmvtalak keniiaaaqa gasgyvvkpf taatleekln
121 kifeklgm
Accession no. 3F7N_A
SOURCE ESCHERIA COLI K-12
AMINO ACID- 128
MAX SCORE – 246
MAX IDENT 98%
1 adkelkflvv ddfstmrriv rnllkelgfn nveeaedgvd alnklqaggf gfiicdwnmp
61 nmdglellkt iradsamsal pvlmvtaeak keniiaaaqa gasgyvvkpf taatleekln
121 kifeklgm
Accession no.- 2CHY_A
Source- Salmonella Enterica Subsp. Eterica Serovar Typhimurium
Amino Acid- 128
Max score- 251
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

BIOINFORMATICS ASSIGNMENT 8
MAX Ident - 97 %
Define the terms score and P value and give example to explain?
E value
The Expectation value or E-value is the parameter which describes the number of hits which can
be expected to see by chance whn finding a database of particular size. When the score of the matches
increase e value decreases exponenetially the lower the E value, the more significant the score and
the alignment.
Example:-
Query: CPn0189 Score E-
(Bits) Value
Aligned with CT131 hypothetical protein 1240 0.0
Query: 1 MKRRSWLKILGICLGSSIVLGFLIFLPQLLSTESRKYLVFSLIHKESGLSCSAEELKISW 60
MKR W KI G L + L L LP+ S+ES KYL S+++KE+GL E+L +SW
Sbjct: 1 MKRSPWYKIFGYYLLVGVPLALLALLPKFFSSESGKYLFLSVLNKETGLQFEIEQLHLSW 60
Query: 61 FGRQTARKIKLTG-EAKDEVFSAEKFELDGSLLRLLIYKKPKGITLSGWSLKINEPASID 119
FG QTA+KI++ G ++ E+F+AEK + GSL RLL+Y+ PK +TL+GWSL+I+E S++
Sbjct: 61 FGSQTAKKIRIRGIDSDSEIFAAEKIIVKGSLPRLLLYRFPKALTLTGWSLQIDESLSMN 120
Figure 1 shows the E value in sequence search by using BLAST
Score
A score is a number which is used to assess the biological relevance of a finding
Figure 2 shows the score (258 bits) in a sequence search
Define the identity and conservative substitution in a protein alignment?
MAX Ident - 97 %
Define the terms score and P value and give example to explain?
E value
The Expectation value or E-value is the parameter which describes the number of hits which can
be expected to see by chance whn finding a database of particular size. When the score of the matches
increase e value decreases exponenetially the lower the E value, the more significant the score and
the alignment.
Example:-
Query: CPn0189 Score E-
(Bits) Value
Aligned with CT131 hypothetical protein 1240 0.0
Query: 1 MKRRSWLKILGICLGSSIVLGFLIFLPQLLSTESRKYLVFSLIHKESGLSCSAEELKISW 60
MKR W KI G L + L L LP+ S+ES KYL S+++KE+GL E+L +SW
Sbjct: 1 MKRSPWYKIFGYYLLVGVPLALLALLPKFFSSESGKYLFLSVLNKETGLQFEIEQLHLSW 60
Query: 61 FGRQTARKIKLTG-EAKDEVFSAEKFELDGSLLRLLIYKKPKGITLSGWSLKINEPASID 119
FG QTA+KI++ G ++ E+F+AEK + GSL RLL+Y+ PK +TL+GWSL+I+E S++
Sbjct: 61 FGSQTAKKIRIRGIDSDSEIFAAEKIIVKGSLPRLLLYRFPKALTLTGWSLQIDESLSMN 120
Figure 1 shows the E value in sequence search by using BLAST
Score
A score is a number which is used to assess the biological relevance of a finding
Figure 2 shows the score (258 bits) in a sequence search
Define the identity and conservative substitution in a protein alignment?

BIOINFORMATICS ASSIGNMENT 9
Identity
Identity is the extents to which two of the sequences have the similar residues at the same
positions in an alignment, often represents as percentage.
Conserved substitution
It is a change at a specific location of an amino acid or less commonly DNA sequence which
preserves the physioco-chemical properties of the real residue or achieves the positive score in a
governing scoring matrix.
What is the default substitution matrix on the BLAST page?
Answer- BLOSUM62 (Block Amino Acid Substitution Matrices 62)
What other matrices are available?
PAM30
PAM70
PAM250
BLOSUM 45
BLOSUM80
BLOSUM62
BLOSUM50
BLOSUM90
List the names for these substitution matrices?
Point accepted mutation 30
Point accepted mutation 70
Point accepted mutation 250
Block Amino Acid Substitution Matrices 80
Block Amino Acid Substitution Matrices 62
Block Amino Acid Substitution Matrices 50
Block Amino Acid Substitution Matrices 90
What is the difference between the main two?
PAM BLOSUM
Identity
Identity is the extents to which two of the sequences have the similar residues at the same
positions in an alignment, often represents as percentage.
Conserved substitution
It is a change at a specific location of an amino acid or less commonly DNA sequence which
preserves the physioco-chemical properties of the real residue or achieves the positive score in a
governing scoring matrix.
What is the default substitution matrix on the BLAST page?
Answer- BLOSUM62 (Block Amino Acid Substitution Matrices 62)
What other matrices are available?
PAM30
PAM70
PAM250
BLOSUM 45
BLOSUM80
BLOSUM62
BLOSUM50
BLOSUM90
List the names for these substitution matrices?
Point accepted mutation 30
Point accepted mutation 70
Point accepted mutation 250
Block Amino Acid Substitution Matrices 80
Block Amino Acid Substitution Matrices 62
Block Amino Acid Substitution Matrices 50
Block Amino Acid Substitution Matrices 90
What is the difference between the main two?
PAM BLOSUM
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

BIOINFORMATICS ASSIGNMENT 10
1. PAM matrices developed to score
alignment between nearly related
protein sequences
BLOSUM matrices can be used to
score alignment between evolutionary
divergent protein sequences
2. Based on global alignment Based on local alignment
3. Alignment in PAM have high
similarity than BLOSUM
alignments
Alignment have low similarity than
PAM alignment
4. Less divergent More divergent
5. Higher numbers in this matrix
naming denotes higher evolutionary
distance
Greater number in the BLOSUM
matrix naming indicates higher
sequence similarity and lesser
evolutionary distance
6. Example: PAM 30, PAM 70 Example: BLOSUM62, BLOSUM 80
Does the use of a different substitution matrix affect the results of the search?
Answer:
By using different substitution matrix (PAM 70) it was found that the e value and scores differs in both
results.
4. Learn To Retrieve Gene/mRNA/Protein Information
A. Use the NCBI database to retrieve information for the 6 genes given below, mRNA and protein
sequence (Learn to use Refseq/Gene/Protein/Nucleotide database)?
Answer
BRCA 2
Gene Type- protein coding
1. PAM matrices developed to score
alignment between nearly related
protein sequences
BLOSUM matrices can be used to
score alignment between evolutionary
divergent protein sequences
2. Based on global alignment Based on local alignment
3. Alignment in PAM have high
similarity than BLOSUM
alignments
Alignment have low similarity than
PAM alignment
4. Less divergent More divergent
5. Higher numbers in this matrix
naming denotes higher evolutionary
distance
Greater number in the BLOSUM
matrix naming indicates higher
sequence similarity and lesser
evolutionary distance
6. Example: PAM 30, PAM 70 Example: BLOSUM62, BLOSUM 80
Does the use of a different substitution matrix affect the results of the search?
Answer:
By using different substitution matrix (PAM 70) it was found that the e value and scores differs in both
results.
4. Learn To Retrieve Gene/mRNA/Protein Information
A. Use the NCBI database to retrieve information for the 6 genes given below, mRNA and protein
sequence (Learn to use Refseq/Gene/Protein/Nucleotide database)?
Answer
BRCA 2
Gene Type- protein coding
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

BIOINFORMATICS ASSIGNMENT 11
Organism –Homo sapiens
Preferred names- breast cancer type 2 susceptibility protein
Function-
It involved in maintenance of genome stability
It considered as the tumour repressor gene
It contains many copies of a 70 aa motif which is called the BRC motif, and these motifs provides
binding to the RAD51 recombinase that functions in DNA repair.
Number of exons present – 27
Location of the gene on the chromosome – 13q13.1 (on chromosome no. 13, NC_000013.11)
Separate protein and mRNA sequence
Accession no. AAG46030
Protein sequence 1
YDTEIDRSRRSAIKKIMERDDTAAKTLVLCVSDIISLSANISETSSSKTSSADTQKVA
Accession no. AAK29432
Protein sequence 2
TAAPKCKEMQNSLNNDKNLVSIETVVPPKLLSDNLCRQTENLKTSKSIFLKVKVHENVEKETAKSPATCY
TNQSPYSVIENSALAFYTSCSRKTSVSQTSLLEAKKWLREGIFDGQPERINTADYVGNYLYENNSNSTIA
ENDKNHLSEKQDTYLSNSSMSNSYSYHSDEVYNDSGYLSKNKLDSGIEPVLKNVEDQKNTSFSKVISNVK
DANAYPQTVNEDICVEELVTSSSPCKNKNAAIKLSISNSNNFEVGPPAFRIASGKIVCVSHETIKKVKDI
FTDSFSKVIKENNENKSKICQTKIMAGCYEALDDSEDILHNSLDNDECSMHSHKVFADIQSEEILQHNQN
MSGLEKVSKISPCDVSLETSDICKCSIGKLHKSVSSANTCGIFSTASGKSVQVSDASLQNARQVFSEIED
STKQVFSKVLFKSNEHSDQLTREENTAIRTPEHLISQKGFSYNVVNSSAFSGFSTASGKQVSILESSLHK
VKGVLEEFDLIRTEHSLHYSPTSRQNVSKILPRVDKRNPEHCVNSEMEKTCSKEFKLSNNLNVEGGSSEN
NHSIKVSPYLSQFQQDKQQLVLGTKVSLVENIHVLGKEQASPKNVKMEIGKTETFSDVPVKTNIEVCSTY
SKDSENYFETEAVEIAKAFMEDDELTDSKLPSHATHSLFTCPENEEMVLSNSRIGKRRGEPLILVGKCSF
LPFVLPITIFKVFIQ
Accession no. AAN61409
Protein sequence 3
MPIGSKERPTFFEIFKTRCNKA
Organism –Homo sapiens
Preferred names- breast cancer type 2 susceptibility protein
Function-
It involved in maintenance of genome stability
It considered as the tumour repressor gene
It contains many copies of a 70 aa motif which is called the BRC motif, and these motifs provides
binding to the RAD51 recombinase that functions in DNA repair.
Number of exons present – 27
Location of the gene on the chromosome – 13q13.1 (on chromosome no. 13, NC_000013.11)
Separate protein and mRNA sequence
Accession no. AAG46030
Protein sequence 1
YDTEIDRSRRSAIKKIMERDDTAAKTLVLCVSDIISLSANISETSSSKTSSADTQKVA
Accession no. AAK29432
Protein sequence 2
TAAPKCKEMQNSLNNDKNLVSIETVVPPKLLSDNLCRQTENLKTSKSIFLKVKVHENVEKETAKSPATCY
TNQSPYSVIENSALAFYTSCSRKTSVSQTSLLEAKKWLREGIFDGQPERINTADYVGNYLYENNSNSTIA
ENDKNHLSEKQDTYLSNSSMSNSYSYHSDEVYNDSGYLSKNKLDSGIEPVLKNVEDQKNTSFSKVISNVK
DANAYPQTVNEDICVEELVTSSSPCKNKNAAIKLSISNSNNFEVGPPAFRIASGKIVCVSHETIKKVKDI
FTDSFSKVIKENNENKSKICQTKIMAGCYEALDDSEDILHNSLDNDECSMHSHKVFADIQSEEILQHNQN
MSGLEKVSKISPCDVSLETSDICKCSIGKLHKSVSSANTCGIFSTASGKSVQVSDASLQNARQVFSEIED
STKQVFSKVLFKSNEHSDQLTREENTAIRTPEHLISQKGFSYNVVNSSAFSGFSTASGKQVSILESSLHK
VKGVLEEFDLIRTEHSLHYSPTSRQNVSKILPRVDKRNPEHCVNSEMEKTCSKEFKLSNNLNVEGGSSEN
NHSIKVSPYLSQFQQDKQQLVLGTKVSLVENIHVLGKEQASPKNVKMEIGKTETFSDVPVKTNIEVCSTY
SKDSENYFETEAVEIAKAFMEDDELTDSKLPSHATHSLFTCPENEEMVLSNSRIGKRRGEPLILVGKCSF
LPFVLPITIFKVFIQ
Accession no. AAN61409
Protein sequence 3
MPIGSKERPTFFEIFKTRCNKA
BIOINFORMATICS ASSIGNMENT 12
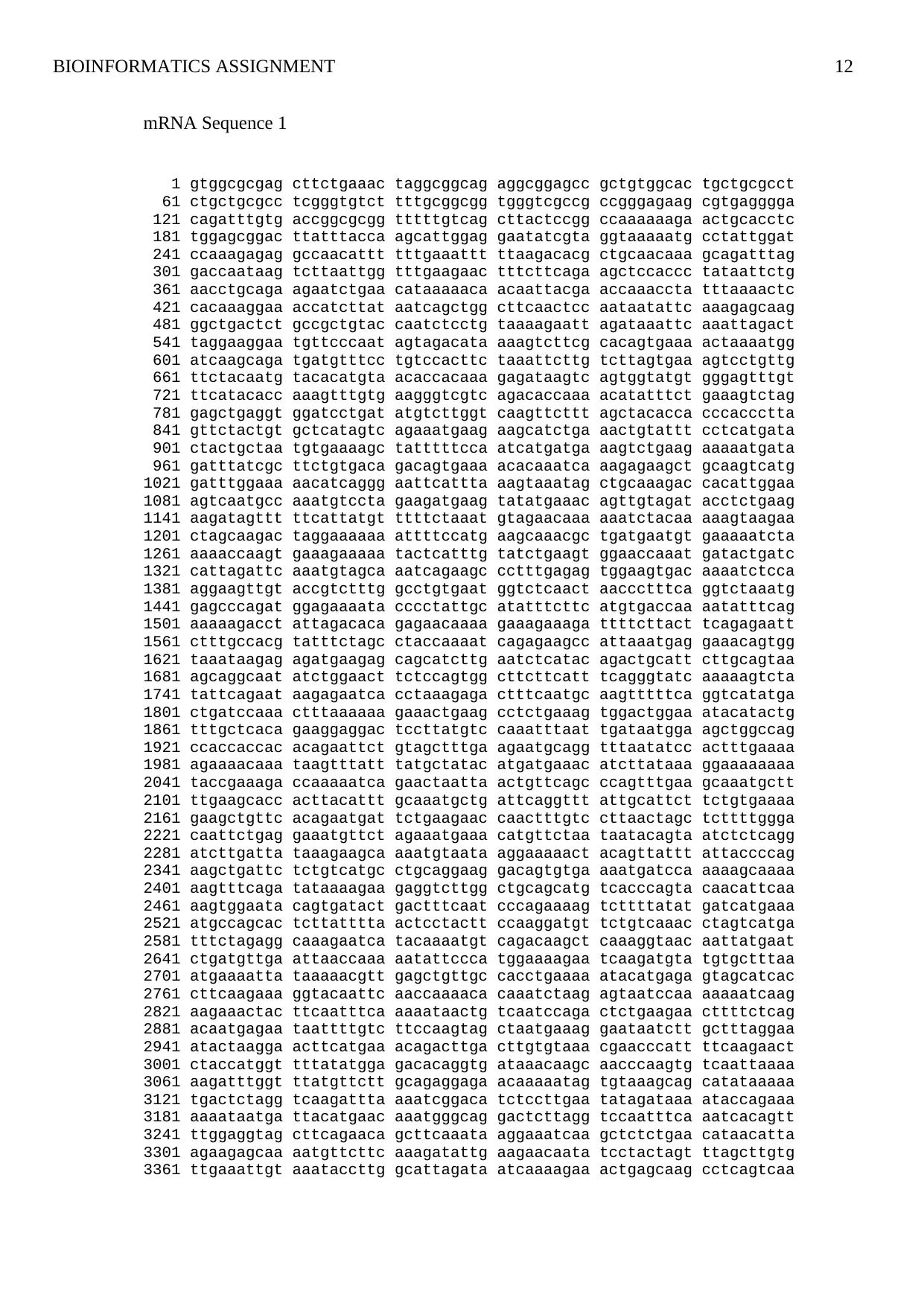
mRNA Sequence 1
1 gtggcgcgag cttctgaaac taggcggcag aggcggagcc gctgtggcac tgctgcgcct
61 ctgctgcgcc tcgggtgtct tttgcggcgg tgggtcgccg ccgggagaag cgtgagggga
121 cagatttgtg accggcgcgg tttttgtcag cttactccgg ccaaaaaaga actgcacctc
181 tggagcggac ttatttacca agcattggag gaatatcgta ggtaaaaatg cctattggat
241 ccaaagagag gccaacattt tttgaaattt ttaagacacg ctgcaacaaa gcagatttag
301 gaccaataag tcttaattgg tttgaagaac tttcttcaga agctccaccc tataattctg
361 aacctgcaga agaatctgaa cataaaaaca acaattacga accaaaccta tttaaaactc
421 cacaaaggaa accatcttat aatcagctgg cttcaactcc aataatattc aaagagcaag
481 ggctgactct gccgctgtac caatctcctg taaaagaatt agataaattc aaattagact
541 taggaaggaa tgttcccaat agtagacata aaagtcttcg cacagtgaaa actaaaatgg
601 atcaagcaga tgatgtttcc tgtccacttc taaattcttg tcttagtgaa agtcctgttg
661 ttctacaatg tacacatgta acaccacaaa gagataagtc agtggtatgt gggagtttgt
721 ttcatacacc aaagtttgtg aagggtcgtc agacaccaaa acatatttct gaaagtctag
781 gagctgaggt ggatcctgat atgtcttggt caagttcttt agctacacca cccaccctta
841 gttctactgt gctcatagtc agaaatgaag aagcatctga aactgtattt cctcatgata
901 ctactgctaa tgtgaaaagc tatttttcca atcatgatga aagtctgaag aaaaatgata
961 gatttatcgc ttctgtgaca gacagtgaaa acacaaatca aagagaagct gcaagtcatg
1021 gatttggaaa aacatcaggg aattcattta aagtaaatag ctgcaaagac cacattggaa
1081 agtcaatgcc aaatgtccta gaagatgaag tatatgaaac agttgtagat acctctgaag
1141 aagatagttt ttcattatgt ttttctaaat gtagaacaaa aaatctacaa aaagtaagaa
1201 ctagcaagac taggaaaaaa attttccatg aagcaaacgc tgatgaatgt gaaaaatcta
1261 aaaaccaagt gaaagaaaaa tactcatttg tatctgaagt ggaaccaaat gatactgatc
1321 cattagattc aaatgtagca aatcagaagc cctttgagag tggaagtgac aaaatctcca
1381 aggaagttgt accgtctttg gcctgtgaat ggtctcaact aaccctttca ggtctaaatg
1441 gagcccagat ggagaaaata cccctattgc atatttcttc atgtgaccaa aatatttcag
1501 aaaaagacct attagacaca gagaacaaaa gaaagaaaga ttttcttact tcagagaatt
1561 ctttgccacg tatttctagc ctaccaaaat cagagaagcc attaaatgag gaaacagtgg
1621 taaataagag agatgaagag cagcatcttg aatctcatac agactgcatt cttgcagtaa
1681 agcaggcaat atctggaact tctccagtgg cttcttcatt tcagggtatc aaaaagtcta
1741 tattcagaat aagagaatca cctaaagaga ctttcaatgc aagtttttca ggtcatatga
1801 ctgatccaaa ctttaaaaaa gaaactgaag cctctgaaag tggactggaa atacatactg
1861 tttgctcaca gaaggaggac tccttatgtc caaatttaat tgataatgga agctggccag
1921 ccaccaccac acagaattct gtagctttga agaatgcagg tttaatatcc actttgaaaa
1981 agaaaacaaa taagtttatt tatgctatac atgatgaaac atcttataaa ggaaaaaaaa
2041 taccgaaaga ccaaaaatca gaactaatta actgttcagc ccagtttgaa gcaaatgctt
2101 ttgaagcacc acttacattt gcaaatgctg attcaggttt attgcattct tctgtgaaaa
2161 gaagctgttc acagaatgat tctgaagaac caactttgtc cttaactagc tcttttggga
2221 caattctgag gaaatgttct agaaatgaaa catgttctaa taatacagta atctctcagg
2281 atcttgatta taaagaagca aaatgtaata aggaaaaact acagttattt attaccccag
2341 aagctgattc tctgtcatgc ctgcaggaag gacagtgtga aaatgatcca aaaagcaaaa
2401 aagtttcaga tataaaagaa gaggtcttgg ctgcagcatg tcacccagta caacattcaa
2461 aagtggaata cagtgatact gactttcaat cccagaaaag tcttttatat gatcatgaaa
2521 atgccagcac tcttatttta actcctactt ccaaggatgt tctgtcaaac ctagtcatga
2581 tttctagagg caaagaatca tacaaaatgt cagacaagct caaaggtaac aattatgaat
2641 ctgatgttga attaaccaaa aatattccca tggaaaagaa tcaagatgta tgtgctttaa
2701 atgaaaatta taaaaacgtt gagctgttgc cacctgaaaa atacatgaga gtagcatcac
2761 cttcaagaaa ggtacaattc aaccaaaaca caaatctaag agtaatccaa aaaaatcaag
2821 aagaaactac ttcaatttca aaaataactg tcaatccaga ctctgaagaa cttttctcag
2881 acaatgagaa taattttgtc ttccaagtag ctaatgaaag gaataatctt gctttaggaa
2941 atactaagga acttcatgaa acagacttga cttgtgtaaa cgaacccatt ttcaagaact
3001 ctaccatggt tttatatgga gacacaggtg ataaacaagc aacccaagtg tcaattaaaa
3061 aagatttggt ttatgttctt gcagaggaga acaaaaatag tgtaaagcag catataaaaa
3121 tgactctagg tcaagattta aaatcggaca tctccttgaa tatagataaa ataccagaaa
3181 aaaataatga ttacatgaac aaatgggcag gactcttagg tccaatttca aatcacagtt
3241 ttggaggtag cttcagaaca gcttcaaata aggaaatcaa gctctctgaa cataacatta
3301 agaagagcaa aatgttcttc aaagatattg aagaacaata tcctactagt ttagcttgtg
3361 ttgaaattgt aaataccttg gcattagata atcaaaagaa actgagcaag cctcagtcaa
mRNA Sequence 1
1 gtggcgcgag cttctgaaac taggcggcag aggcggagcc gctgtggcac tgctgcgcct
61 ctgctgcgcc tcgggtgtct tttgcggcgg tgggtcgccg ccgggagaag cgtgagggga
121 cagatttgtg accggcgcgg tttttgtcag cttactccgg ccaaaaaaga actgcacctc
181 tggagcggac ttatttacca agcattggag gaatatcgta ggtaaaaatg cctattggat
241 ccaaagagag gccaacattt tttgaaattt ttaagacacg ctgcaacaaa gcagatttag
301 gaccaataag tcttaattgg tttgaagaac tttcttcaga agctccaccc tataattctg
361 aacctgcaga agaatctgaa cataaaaaca acaattacga accaaaccta tttaaaactc
421 cacaaaggaa accatcttat aatcagctgg cttcaactcc aataatattc aaagagcaag
481 ggctgactct gccgctgtac caatctcctg taaaagaatt agataaattc aaattagact
541 taggaaggaa tgttcccaat agtagacata aaagtcttcg cacagtgaaa actaaaatgg
601 atcaagcaga tgatgtttcc tgtccacttc taaattcttg tcttagtgaa agtcctgttg
661 ttctacaatg tacacatgta acaccacaaa gagataagtc agtggtatgt gggagtttgt
721 ttcatacacc aaagtttgtg aagggtcgtc agacaccaaa acatatttct gaaagtctag
781 gagctgaggt ggatcctgat atgtcttggt caagttcttt agctacacca cccaccctta
841 gttctactgt gctcatagtc agaaatgaag aagcatctga aactgtattt cctcatgata
901 ctactgctaa tgtgaaaagc tatttttcca atcatgatga aagtctgaag aaaaatgata
961 gatttatcgc ttctgtgaca gacagtgaaa acacaaatca aagagaagct gcaagtcatg
1021 gatttggaaa aacatcaggg aattcattta aagtaaatag ctgcaaagac cacattggaa
1081 agtcaatgcc aaatgtccta gaagatgaag tatatgaaac agttgtagat acctctgaag
1141 aagatagttt ttcattatgt ttttctaaat gtagaacaaa aaatctacaa aaagtaagaa
1201 ctagcaagac taggaaaaaa attttccatg aagcaaacgc tgatgaatgt gaaaaatcta
1261 aaaaccaagt gaaagaaaaa tactcatttg tatctgaagt ggaaccaaat gatactgatc
1321 cattagattc aaatgtagca aatcagaagc cctttgagag tggaagtgac aaaatctcca
1381 aggaagttgt accgtctttg gcctgtgaat ggtctcaact aaccctttca ggtctaaatg
1441 gagcccagat ggagaaaata cccctattgc atatttcttc atgtgaccaa aatatttcag
1501 aaaaagacct attagacaca gagaacaaaa gaaagaaaga ttttcttact tcagagaatt
1561 ctttgccacg tatttctagc ctaccaaaat cagagaagcc attaaatgag gaaacagtgg
1621 taaataagag agatgaagag cagcatcttg aatctcatac agactgcatt cttgcagtaa
1681 agcaggcaat atctggaact tctccagtgg cttcttcatt tcagggtatc aaaaagtcta
1741 tattcagaat aagagaatca cctaaagaga ctttcaatgc aagtttttca ggtcatatga
1801 ctgatccaaa ctttaaaaaa gaaactgaag cctctgaaag tggactggaa atacatactg
1861 tttgctcaca gaaggaggac tccttatgtc caaatttaat tgataatgga agctggccag
1921 ccaccaccac acagaattct gtagctttga agaatgcagg tttaatatcc actttgaaaa
1981 agaaaacaaa taagtttatt tatgctatac atgatgaaac atcttataaa ggaaaaaaaa
2041 taccgaaaga ccaaaaatca gaactaatta actgttcagc ccagtttgaa gcaaatgctt
2101 ttgaagcacc acttacattt gcaaatgctg attcaggttt attgcattct tctgtgaaaa
2161 gaagctgttc acagaatgat tctgaagaac caactttgtc cttaactagc tcttttggga
2221 caattctgag gaaatgttct agaaatgaaa catgttctaa taatacagta atctctcagg
2281 atcttgatta taaagaagca aaatgtaata aggaaaaact acagttattt attaccccag
2341 aagctgattc tctgtcatgc ctgcaggaag gacagtgtga aaatgatcca aaaagcaaaa
2401 aagtttcaga tataaaagaa gaggtcttgg ctgcagcatg tcacccagta caacattcaa
2461 aagtggaata cagtgatact gactttcaat cccagaaaag tcttttatat gatcatgaaa
2521 atgccagcac tcttatttta actcctactt ccaaggatgt tctgtcaaac ctagtcatga
2581 tttctagagg caaagaatca tacaaaatgt cagacaagct caaaggtaac aattatgaat
2641 ctgatgttga attaaccaaa aatattccca tggaaaagaa tcaagatgta tgtgctttaa
2701 atgaaaatta taaaaacgtt gagctgttgc cacctgaaaa atacatgaga gtagcatcac
2761 cttcaagaaa ggtacaattc aaccaaaaca caaatctaag agtaatccaa aaaaatcaag
2821 aagaaactac ttcaatttca aaaataactg tcaatccaga ctctgaagaa cttttctcag
2881 acaatgagaa taattttgtc ttccaagtag ctaatgaaag gaataatctt gctttaggaa
2941 atactaagga acttcatgaa acagacttga cttgtgtaaa cgaacccatt ttcaagaact
3001 ctaccatggt tttatatgga gacacaggtg ataaacaagc aacccaagtg tcaattaaaa
3061 aagatttggt ttatgttctt gcagaggaga acaaaaatag tgtaaagcag catataaaaa
3121 tgactctagg tcaagattta aaatcggaca tctccttgaa tatagataaa ataccagaaa
3181 aaaataatga ttacatgaac aaatgggcag gactcttagg tccaatttca aatcacagtt
3241 ttggaggtag cttcagaaca gcttcaaata aggaaatcaa gctctctgaa cataacatta
3301 agaagagcaa aatgttcttc aaagatattg aagaacaata tcctactagt ttagcttgtg
3361 ttgaaattgt aaataccttg gcattagata atcaaaagaa actgagcaag cctcagtcaa
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 56
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.





