Bioinformatics Report: Phylogenetic Prediction and Analysis
VerifiedAdded on 2022/07/21
|5
|1299
|21
Report
AI Summary
This report provides an overview of phylogenetic prediction, a crucial aspect of bioinformatics used to determine evolutionary relationships between sequences. It explores various methods for constructing evolutionary trees, including maximum parsimony, distance methods, and maximum likelihood. The report emphasizes the importance of multiple sequence alignment (MSA) and the factors to consider when selecting a suitable phylogenetic method. It explains how maximum parsimony identifies the tree with the fewest evolutionary steps, while also touching upon distance-based methods. The report also discusses the limitations of each method, such as the computational intensity of maximum likelihood and the need for sequence similarity in maximum parsimony. Overall, the document aims to guide researchers in selecting and applying the appropriate phylogenetic prediction methods for analyzing a set of related sequences.

Contents
Phylogenetic prediction.............................................................................................................................3
1. Phylogenetic......................................................................................................................................3
1.1. Methods for determining evolutionary trees............................................................................3
1.1.1. Maximum Parsimony.........................................................................................................5
Table of Figure:
Figure 10 The Point considering while selecting a method.......................................................................3
Figure 11 Multiple sequence alignment....................................................................................................5
Phylogenetic prediction.............................................................................................................................3
1. Phylogenetic......................................................................................................................................3
1.1. Methods for determining evolutionary trees............................................................................3
1.1.1. Maximum Parsimony.........................................................................................................5
Table of Figure:
Figure 10 The Point considering while selecting a method.......................................................................3
Figure 11 Multiple sequence alignment....................................................................................................5
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Phylogenetic prediction
1. Phylogenetic
1.1. Methods for determining evolutionary trees
There are three ways to find the tree (or trees) that best explain the differences seen in a set of
sequences. These ways are called "maximum likelihood," "maximum parsimony," and "maximum
distance," respectively. When deciding which of these methods to use, we should consider the
sequences you want to compare. When there is a lot of overlap between the sequences,
maximum parsimony is the best way. Distance methods work best when there isn't a lot of
similarity between two sequences, but there is some similarity that is easy to spot. The
method to use for everything else is the maximum likelihood model.
Researchers usually use one of three methods (maximum parsimony, distance, or maximum
likelihood) to find the evolutionary tree or trees that make the most sense for the differences
they see in sequences. One of these approaches utilizes a different method of analysis than
the others. Distance-based programs are often used in molecular biology because they are
easy to use and can be used on many sequences. Methods that use the maximum likelihood
are harder to use and require a better understanding of the evolutionary models they are
based on. Because maximum likelihood programs are so computer-intensive and the number
of steps needed to analyze each sequence grows exponentially with the number of sequences
being analyzed, they can only look at a certain number of sequences. They can be run on a
supercomputer, which lets more sequences be looked at. This article gives a summary for a
researcher who has a set of related sequences and wants to figure out the best way to show
the phylogenetic relationships between the sequences by making a tree. This article is written
for a researcher who wants to look at a set of related sequences.
1. Phylogenetic
1.1. Methods for determining evolutionary trees
There are three ways to find the tree (or trees) that best explain the differences seen in a set of
sequences. These ways are called "maximum likelihood," "maximum parsimony," and "maximum
distance," respectively. When deciding which of these methods to use, we should consider the
sequences you want to compare. When there is a lot of overlap between the sequences,
maximum parsimony is the best way. Distance methods work best when there isn't a lot of
similarity between two sequences, but there is some similarity that is easy to spot. The
method to use for everything else is the maximum likelihood model.
Researchers usually use one of three methods (maximum parsimony, distance, or maximum
likelihood) to find the evolutionary tree or trees that make the most sense for the differences
they see in sequences. One of these approaches utilizes a different method of analysis than
the others. Distance-based programs are often used in molecular biology because they are
easy to use and can be used on many sequences. Methods that use the maximum likelihood
are harder to use and require a better understanding of the evolutionary models they are
based on. Because maximum likelihood programs are so computer-intensive and the number
of steps needed to analyze each sequence grows exponentially with the number of sequences
being analyzed, they can only look at a certain number of sequences. They can be run on a
supercomputer, which lets more sequences be looked at. This article gives a summary for a
researcher who has a set of related sequences and wants to figure out the best way to show
the phylogenetic relationships between the sequences by making a tree. This article is written
for a researcher who wants to look at a set of related sequences.
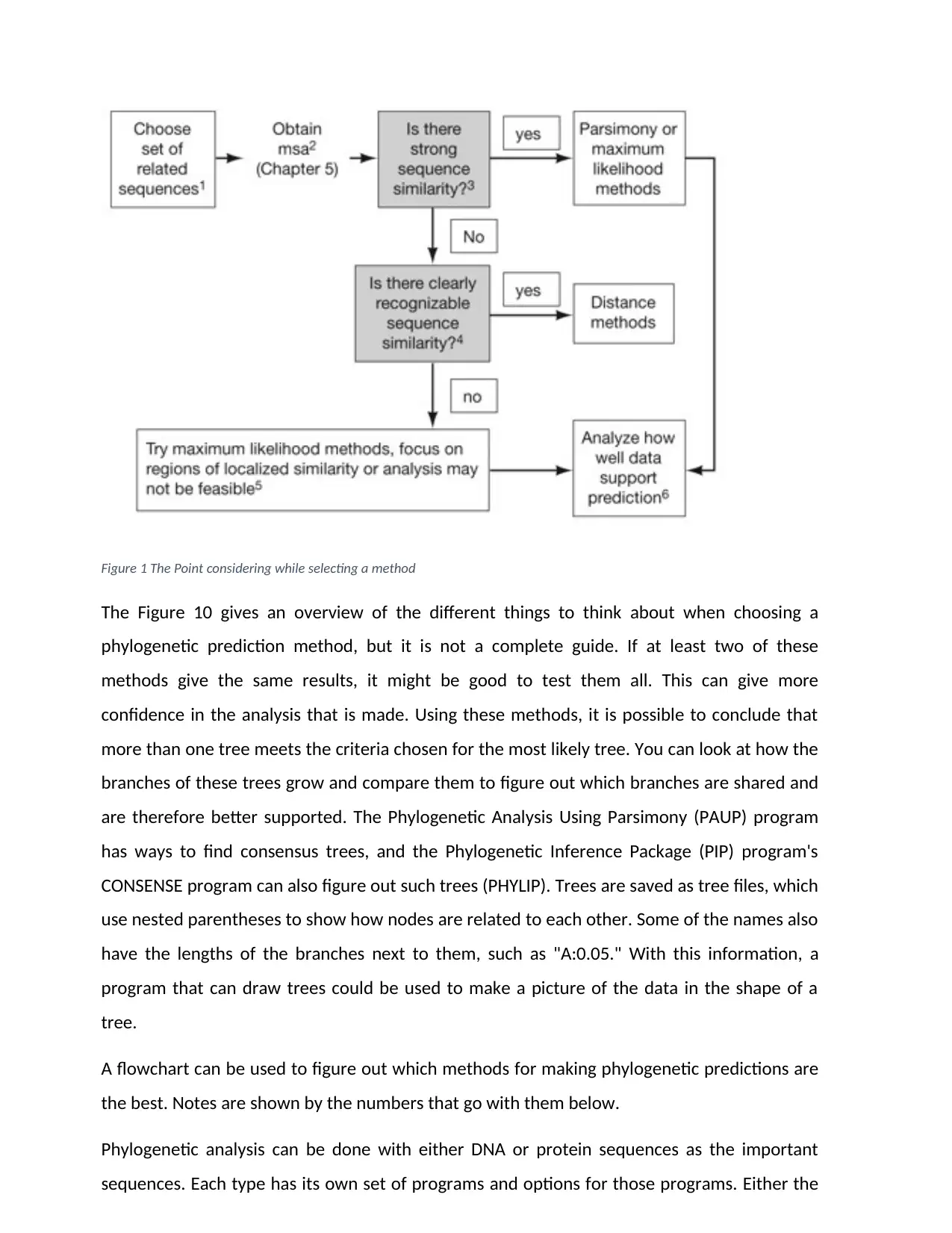
Figure 1 The Point considering while selecting a method
The Figure 10 gives an overview of the different things to think about when choosing a
phylogenetic prediction method, but it is not a complete guide. If at least two of these
methods give the same results, it might be good to test them all. This can give more
confidence in the analysis that is made. Using these methods, it is possible to conclude that
more than one tree meets the criteria chosen for the most likely tree. You can look at how the
branches of these trees grow and compare them to figure out which branches are shared and
are therefore better supported. The Phylogenetic Analysis Using Parsimony (PAUP) program
has ways to find consensus trees, and the Phylogenetic Inference Package (PIP) program's
CONSENSE program can also figure out such trees (PHYLIP). Trees are saved as tree files, which
use nested parentheses to show how nodes are related to each other. Some of the names also
have the lengths of the branches next to them, such as "A:0.05." With this information, a
program that can draw trees could be used to make a picture of the data in the shape of a
tree.
A flowchart can be used to figure out which methods for making phylogenetic predictions are
the best. Notes are shown by the numbers that go with them below.
Phylogenetic analysis can be done with either DNA or protein sequences as the important
sequences. Each type has its own set of programs and options for those programs. Either the
The Figure 10 gives an overview of the different things to think about when choosing a
phylogenetic prediction method, but it is not a complete guide. If at least two of these
methods give the same results, it might be good to test them all. This can give more
confidence in the analysis that is made. Using these methods, it is possible to conclude that
more than one tree meets the criteria chosen for the most likely tree. You can look at how the
branches of these trees grow and compare them to figure out which branches are shared and
are therefore better supported. The Phylogenetic Analysis Using Parsimony (PAUP) program
has ways to find consensus trees, and the Phylogenetic Inference Package (PIP) program's
CONSENSE program can also figure out such trees (PHYLIP). Trees are saved as tree files, which
use nested parentheses to show how nodes are related to each other. Some of the names also
have the lengths of the branches next to them, such as "A:0.05." With this information, a
program that can draw trees could be used to make a picture of the data in the shape of a
tree.
A flowchart can be used to figure out which methods for making phylogenetic predictions are
the best. Notes are shown by the numbers that go with them below.
Phylogenetic analysis can be done with either DNA or protein sequences as the important
sequences. Each type has its own set of programs and options for those programs. Either the
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

sequences should match up along their whole lengths or share a set of patterns or domains
that is a strong sign that they evolved together. If this isn't the case, the sequences shouldn't
have anything to do with each other. Analysis of RNA sequences is done with the help of
covariation and changes in secondary structure.
Based on the genetic distances between sequence pairs in a multiple sequence alignment
(MSA), distance methods can be used to analyze an MSA's phylogenetic analysis. This makes it
possible to do the analysis. The genetic distance between two sequences is the number of
places along an aligned sequence where one sequence is different from the other. On the
other hand, sequence identity is a measure of how many aligned positions are the same from
one sequence to the next. Gaps can be ignored or treated as if they were substitutions when
figuring out how far something is. You can also use a scoring matrix or a substitution matrix,
which makes the calculation more complicated but doesn't change the basic idea. "Neighbors"
are the relationships between sequence pairs close to each other. These sequences are set up
on a tree so that their separate branches all lead to the same node, which is their common
ancestor.
1.1.1. Maximum Parsimony
The maximum parsimony method predicts the evolutionary tree or trees that need the fewest
steps to explain how the observed differences in the sequences come from the same ancestral
sequences. The method is also sometimes called the method of minimum evolution. A
multiple sequence alignment (MSA) is needed to figure out which parts of two sequences are
most likely to match each other. The MSA will show these positions as columns going up and
down. The phylogenetic trees that need the fewest number of changes to explain the
observed changes in sequences from ancestral sequences are found for each aligned position.
These trees cause the observed changes in the ancestral sequences. For each position in the
sequence alignment, the analysis is done. The trees are put in order based on which ones
make the fewest changes for every position in the sequence. This method works best with
very similar sequences, and it can only be used on a small number of sequences at once.
Maximum parsimony methods predict the evolutionary tree with the fewest steps needed to
explain the differences in the sequences. The principle of maximum parsimony says that you
must first get a multiple sequence alignment to build a tree. For each position that has been
that is a strong sign that they evolved together. If this isn't the case, the sequences shouldn't
have anything to do with each other. Analysis of RNA sequences is done with the help of
covariation and changes in secondary structure.
Based on the genetic distances between sequence pairs in a multiple sequence alignment
(MSA), distance methods can be used to analyze an MSA's phylogenetic analysis. This makes it
possible to do the analysis. The genetic distance between two sequences is the number of
places along an aligned sequence where one sequence is different from the other. On the
other hand, sequence identity is a measure of how many aligned positions are the same from
one sequence to the next. Gaps can be ignored or treated as if they were substitutions when
figuring out how far something is. You can also use a scoring matrix or a substitution matrix,
which makes the calculation more complicated but doesn't change the basic idea. "Neighbors"
are the relationships between sequence pairs close to each other. These sequences are set up
on a tree so that their separate branches all lead to the same node, which is their common
ancestor.
1.1.1. Maximum Parsimony
The maximum parsimony method predicts the evolutionary tree or trees that need the fewest
steps to explain how the observed differences in the sequences come from the same ancestral
sequences. The method is also sometimes called the method of minimum evolution. A
multiple sequence alignment (MSA) is needed to figure out which parts of two sequences are
most likely to match each other. The MSA will show these positions as columns going up and
down. The phylogenetic trees that need the fewest number of changes to explain the
observed changes in sequences from ancestral sequences are found for each aligned position.
These trees cause the observed changes in the ancestral sequences. For each position in the
sequence alignment, the analysis is done. The trees are put in order based on which ones
make the fewest changes for every position in the sequence. This method works best with
very similar sequences, and it can only be used on a small number of sequences at once.
Maximum parsimony methods predict the evolutionary tree with the fewest steps needed to
explain the differences in the sequences. The principle of maximum parsimony says that you
must first get a multiple sequence alignment to build a tree. For each position that has been
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

aligned, it is possible to find the phylogenetic trees that need the fewest changes to explain
the sequence differences found. For each position in the alignment, the steps above are done
again. Those trees are chosen for identification with the fewest number of changes in each
sequence position overall. This is a task that takes a lot of time. The algorithm takes a lot of
time and can only work if the sequences are very similar as shown in figure 10.
Figure 2 Multiple sequence alignment
Take the previous example into account (Mount, 250). There are four sequences, which means
that three different trees could grow but haven't started yet. In this case, some websites are
helpful, and others are not helpful. At least two different sequences on an informative website
will have the same sequence character in the same place. Only the sites that have useful
information should be taken into account.
the sequence differences found. For each position in the alignment, the steps above are done
again. Those trees are chosen for identification with the fewest number of changes in each
sequence position overall. This is a task that takes a lot of time. The algorithm takes a lot of
time and can only work if the sequences are very similar as shown in figure 10.
Figure 2 Multiple sequence alignment
Take the previous example into account (Mount, 250). There are four sequences, which means
that three different trees could grow but haven't started yet. In this case, some websites are
helpful, and others are not helpful. At least two different sequences on an informative website
will have the same sequence character in the same place. Only the sites that have useful
information should be taken into account.
1 out of 5
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.





