University Biology Assignment: Genomics and Sequence Analysis
VerifiedAdded on 2022/09/06
|8
|1292
|19
Homework Assignment
AI Summary
This biology assignment delves into the realm of genomics, encompassing topics such as DNA sequencing, multiple sequence alignment (MSA), and gene expression regulation. It begins by comparing and contrasting the services offered by genetic testing companies like 23andMe and Veritas, highlighting the differences in their methodologies and pricing structures. The assignment then explores MSA, explaining its function and the interpretation of conserved residues within aligned sequences. The subsequent sections focus on protein motifs, specifically the P loop or Walker A motif, including its function, conserved residues, and the impact of mutations. Finally, the assignment examines gene expression, covering translation, RNA splicing, and the role of circular RNAs in transcriptome regulation. The solution provides detailed answers to each task, along with relevant diagrams and a comprehensive bibliography.

Running head: BIOLOGY
GENOMICS
Name of the Student
Name of the University
Author Note
GENOMICS
Name of the Student
Name of the University
Author Note
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

1BIOLOGY
Table of Contents
Task 1.........................................................................................................................................2
Task 2.........................................................................................................................................4
Answer i.................................................................................................................................4
Answer ii................................................................................................................................4
Answer iii...............................................................................................................................4
Answer iv...............................................................................................................................5
Answer v................................................................................................................................5
Task 3.........................................................................................................................................5
Answer 1................................................................................................................................5
Answer 2................................................................................................................................5
Bibliography...............................................................................................................................6
Table of Contents
Task 1.........................................................................................................................................2
Task 2.........................................................................................................................................4
Answer i.................................................................................................................................4
Answer ii................................................................................................................................4
Answer iii...............................................................................................................................4
Answer iv...............................................................................................................................5
Answer v................................................................................................................................5
Task 3.........................................................................................................................................5
Answer 1................................................................................................................................5
Answer 2................................................................................................................................5
Bibliography...............................................................................................................................6

2BIOLOGY
Task 1
Answer 1
23 and Me is the term used to describe the 23 pairs of chromosome in the human DNA.
Ancestry estimates with the saliva samples are estimated up to 0.1% sensitivity. This
organization has made it possible to find relatives across both the globes and streets. 23 and
Me ancestry features have been found to help in searching the historical records associated
with death, birth and marriage certificates. 23 and Me has been found to successfully trace
the lineage and analyze the DNA traced to a lineage. The 23 and Me database has been found
to consist of more than five million of genotyped customers on a worldwide basis. As the
database grows, it has been stated by the company that new relatives will be found. This
company has been found to provide a kit for USD$99 for ancestry identification.
On the other hand, Veritas is a rival company for whole-genome sequencing which states that
23 and Me provides limited genetic information based on just a fraction of DNA. Veritas
does a whole genome sequencing and the cost of the kit is higher than 23 and Me. The cost
amounts to %$599 which gives more information apart from just the ancestry details given by
23 and Me. The genomic database for this company is much higher than that of 23 and Me.
23 and Me just works with 0.1% of the DNA whereas Veritas analyzes the whole genome
sequences. The whole-genome sequencing data has been found to provide many insights on
various genetic diseases which cannot be done by the sequencing data obtained from 23 and
Me.
Thus, on comparing both, it can be stated that although both the organisms are well known
for DNA sequencing, Veritas has a higher cost for the kits than 23 and Me because the former
is engaged in whole-genome sequencing.
Answer 2
Task 1
Answer 1
23 and Me is the term used to describe the 23 pairs of chromosome in the human DNA.
Ancestry estimates with the saliva samples are estimated up to 0.1% sensitivity. This
organization has made it possible to find relatives across both the globes and streets. 23 and
Me ancestry features have been found to help in searching the historical records associated
with death, birth and marriage certificates. 23 and Me has been found to successfully trace
the lineage and analyze the DNA traced to a lineage. The 23 and Me database has been found
to consist of more than five million of genotyped customers on a worldwide basis. As the
database grows, it has been stated by the company that new relatives will be found. This
company has been found to provide a kit for USD$99 for ancestry identification.
On the other hand, Veritas is a rival company for whole-genome sequencing which states that
23 and Me provides limited genetic information based on just a fraction of DNA. Veritas
does a whole genome sequencing and the cost of the kit is higher than 23 and Me. The cost
amounts to %$599 which gives more information apart from just the ancestry details given by
23 and Me. The genomic database for this company is much higher than that of 23 and Me.
23 and Me just works with 0.1% of the DNA whereas Veritas analyzes the whole genome
sequences. The whole-genome sequencing data has been found to provide many insights on
various genetic diseases which cannot be done by the sequencing data obtained from 23 and
Me.
Thus, on comparing both, it can be stated that although both the organisms are well known
for DNA sequencing, Veritas has a higher cost for the kits than 23 and Me because the former
is engaged in whole-genome sequencing.
Answer 2
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

3BIOLOGY
MSA is also known as multiple sequence alignment. This means that a large number of
genomic sequences are aligned together inside a software (online or offline) in order to check
their identity, similarity of non-identity. Clustal W and omega are similar MSA programs
which use seeded guide trees and HMM profiles techniques for the generation of alignments
for three or more sequences. In the alignment figures, some of the sequence groups are
termed as conserved sequences. This is because of the fact that conserved sequences do not
change or vary between one organism to another. A conserved sequence is defined as a
sequence that is present in the genome despite the forces of mutation or species features
which changes the genomic sequences between organisms. These sequences have been found
to occur in both coding and non-coding sequences of nucleic acids. Conserved sequences
originate from the mutations in the highly conserved regions which leads to the generation of
non-viable life forms which gets eliminated via natural selection.
MSA is also known as multiple sequence alignment. This means that a large number of
genomic sequences are aligned together inside a software (online or offline) in order to check
their identity, similarity of non-identity. Clustal W and omega are similar MSA programs
which use seeded guide trees and HMM profiles techniques for the generation of alignments
for three or more sequences. In the alignment figures, some of the sequence groups are
termed as conserved sequences. This is because of the fact that conserved sequences do not
change or vary between one organism to another. A conserved sequence is defined as a
sequence that is present in the genome despite the forces of mutation or species features
which changes the genomic sequences between organisms. These sequences have been found
to occur in both coding and non-coding sequences of nucleic acids. Conserved sequences
originate from the mutations in the highly conserved regions which leads to the generation of
non-viable life forms which gets eliminated via natural selection.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

4BIOLOGY
Task 2
Answer i.
P loop motif is a functional characteristic of protein superfamilies which diverges
from a common ancestor. Various ATP binding domains have been found to have a same less
conserved site with the same function associated with the P loop. The most significant
function of Walker A P loop is the binding of NTPase enzymes and the mediation of terminal
phosphate groups NTPs transfer.
Answer ii.
The most conserved residue of the P loop protein is lysine (K). This residue is very
essential to the conformation of the canonical P loop and is also responsible for directly
interacting with beta and gamma phosphates of the bound NTP.
Answer iii.
The codon for lysine is TTT. Now, to induce a mutation without any activity change
of the protein, TTT needs to be changed to a different codon so than lysine does not coded
However, the coded amino acid must be charged since lysine is a positively charged amino
acid. Lysine can be changed to arginine with a codon of CGG. Thus the new sequence must
be-
5’-NNNNTTTNNNN-3’to 5’-NNNNCGGNNNN-3’
Thus, the primer required for the synthesis of the inactive mutant gene is:
3’NNNNCCGNNNN-5’_ Forward primer
Task 2
Answer i.
P loop motif is a functional characteristic of protein superfamilies which diverges
from a common ancestor. Various ATP binding domains have been found to have a same less
conserved site with the same function associated with the P loop. The most significant
function of Walker A P loop is the binding of NTPase enzymes and the mediation of terminal
phosphate groups NTPs transfer.
Answer ii.
The most conserved residue of the P loop protein is lysine (K). This residue is very
essential to the conformation of the canonical P loop and is also responsible for directly
interacting with beta and gamma phosphates of the bound NTP.
Answer iii.
The codon for lysine is TTT. Now, to induce a mutation without any activity change
of the protein, TTT needs to be changed to a different codon so than lysine does not coded
However, the coded amino acid must be charged since lysine is a positively charged amino
acid. Lysine can be changed to arginine with a codon of CGG. Thus the new sequence must
be-
5’-NNNNTTTNNNN-3’to 5’-NNNNCGGNNNN-3’
Thus, the primer required for the synthesis of the inactive mutant gene is:
3’NNNNCCGNNNN-5’_ Forward primer

5BIOLOGY
Answer iv.
The above stated primer has only one significant feature. This feature states that it
will synthesize only a positively charged amino acid (arginine) to replace a similar positively
charged amino acid (lysine).
Answer v.
Fig 1: Flow chart of mutagenesis process
Task 3
Answer 1
Translation sequence:
5’ATGTCGTAGCTAGATTTGCAGGTCGATGAAGCTTGCTGTTCAGGTAAGTCTGTT
TGTAGGTCACTTGCTTTGC AGGTTTTTGTAAGTGA 3’
Pre-mRNA sequence:
Answer iv.
The above stated primer has only one significant feature. This feature states that it
will synthesize only a positively charged amino acid (arginine) to replace a similar positively
charged amino acid (lysine).
Answer v.
Fig 1: Flow chart of mutagenesis process
Task 3
Answer 1
Translation sequence:
5’ATGTCGTAGCTAGATTTGCAGGTCGATGAAGCTTGCTGTTCAGGTAAGTCTGTT
TGTAGGTCACTTGCTTTGC AGGTTTTTGTAAGTGA 3’
Pre-mRNA sequence:
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
6BIOLOGY
5’AUGUCGUGCUAGAUUUGCAGGUCGAUGAAGCUUGCUGUUCAGGUAAGUCUG
UUUGUAGGUCACUUGCUUUGCAGGUUUUUGUAAGUGA 3’
(i) Translation will start from ATG codon because it gets converted to AUG or the
methionine coding sequence which is the initiating sequence for translation.
(ii) RNA splicing is responsible for giving rise to a mature RNA. In this case, mature mRNA
will be produced which will be translated. Thus, regulation of RNA splicing will also regulate
the beginning of regulation from the initiator codon (AUG or ATG).
(iii) Amino acid sequence:
NT end- MS*LDLQVDEACCSGKSVCRSLALQVFVSX- CT end
(iv) The sequence will be same, but circular in nature.
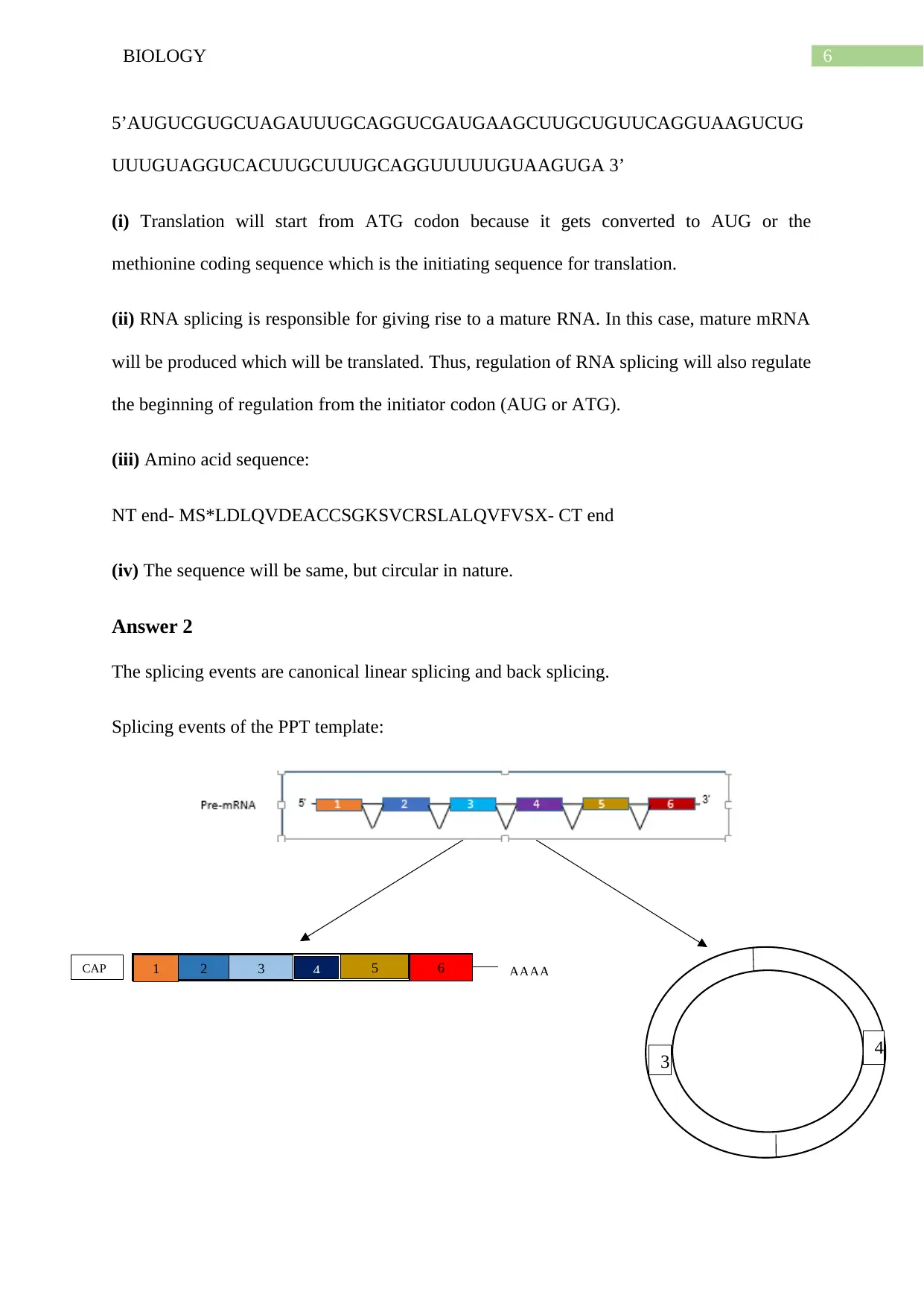
Answer 2
The splicing events are canonical linear splicing and back splicing.
Splicing events of the PPT template:
1 2 3 4 5 6 AAAACAP
3 4
5’AUGUCGUGCUAGAUUUGCAGGUCGAUGAAGCUUGCUGUUCAGGUAAGUCUG
UUUGUAGGUCACUUGCUUUGCAGGUUUUUGUAAGUGA 3’
(i) Translation will start from ATG codon because it gets converted to AUG or the
methionine coding sequence which is the initiating sequence for translation.
(ii) RNA splicing is responsible for giving rise to a mature RNA. In this case, mature mRNA
will be produced which will be translated. Thus, regulation of RNA splicing will also regulate
the beginning of regulation from the initiator codon (AUG or ATG).
(iii) Amino acid sequence:
NT end- MS*LDLQVDEACCSGKSVCRSLALQVFVSX- CT end
(iv) The sequence will be same, but circular in nature.
Answer 2
The splicing events are canonical linear splicing and back splicing.
Splicing events of the PPT template:
1 2 3 4 5 6 AAAACAP
3 4
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

7BIOLOGY
Bibliography
23andme.com. 2020. DNA Genetic Testing & Analysis - 23Andme UK. [online] Available at:
<https://www.23andme.com/?staticrtb=true&flag=true> [Accessed 3 April 2020].
de Opakua, A.I., Parag-Sharma, K., DiGiacomo, V., Merino, N., Leyme, A., Marivin, A.,
Villate, M., Nguyen, L.T., de La Cruz-Morcillo, M.A., Blanco-Canosa, J.B. and
Ramachandran, S., 2017. Molecular mechanism of Gαi activation by non-GPCR proteins
with a Gα-Binding and Activating motif. Nature communications, 8, p.15163.
Ebi.ac.uk. 2020. Clustal Omega < Multiple Sequence Alignment < EMBL-EBI. [online]
Available at: <https://www.ebi.ac.uk/Tools/msa/clustalo/> [Accessed 3 April 2020].
EMBOSS translation tool
ExPASy tool
Huang, S., Yang, B., Chen, B.J., Bliim, N., Ueberham, U., Arendt, T. and Janitz, M., 2017.
The emerging role of circular RNAs in transcriptome regulation. Genomics, 109(5-6),
pp.401-407.
Veritasgenetics.com. 2020. Whole Genome Sequencing And Interpretation | Mygenome |
Veritas Genetics. [online] Available at: <https://www.veritasgenetics.com/myGenome>
[Accessed 3 April 2020].
Yeo, B.P.H., Foong, L.C., Tam, S.M., Lee, V. and Hwang, S.S., 2018. Gene isolation using
degenerate primers targeting protein motif: A laboratory exercise. Biochemistry and
Molecular Biology Education, 46(1), pp.47-53.
Bibliography
23andme.com. 2020. DNA Genetic Testing & Analysis - 23Andme UK. [online] Available at:
<https://www.23andme.com/?staticrtb=true&flag=true> [Accessed 3 April 2020].
de Opakua, A.I., Parag-Sharma, K., DiGiacomo, V., Merino, N., Leyme, A., Marivin, A.,
Villate, M., Nguyen, L.T., de La Cruz-Morcillo, M.A., Blanco-Canosa, J.B. and
Ramachandran, S., 2017. Molecular mechanism of Gαi activation by non-GPCR proteins
with a Gα-Binding and Activating motif. Nature communications, 8, p.15163.
Ebi.ac.uk. 2020. Clustal Omega < Multiple Sequence Alignment < EMBL-EBI. [online]
Available at: <https://www.ebi.ac.uk/Tools/msa/clustalo/> [Accessed 3 April 2020].
EMBOSS translation tool
ExPASy tool
Huang, S., Yang, B., Chen, B.J., Bliim, N., Ueberham, U., Arendt, T. and Janitz, M., 2017.
The emerging role of circular RNAs in transcriptome regulation. Genomics, 109(5-6),
pp.401-407.
Veritasgenetics.com. 2020. Whole Genome Sequencing And Interpretation | Mygenome |
Veritas Genetics. [online] Available at: <https://www.veritasgenetics.com/myGenome>
[Accessed 3 April 2020].
Yeo, B.P.H., Foong, L.C., Tam, S.M., Lee, V. and Hwang, S.S., 2018. Gene isolation using
degenerate primers targeting protein motif: A laboratory exercise. Biochemistry and
Molecular Biology Education, 46(1), pp.47-53.
1 out of 8
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.