BIOL 101: University Bradford Protein Assay Practical Report
VerifiedAdded on 2020/05/16
|9
|2033
|151
Practical Assignment
AI Summary
This document details a biology practical assignment focused on the Bradford Protein Assay, a colorimetric method used to determine protein concentration in a sample. The experiment involves preparing DCPIP solutions, measuring absorbance using a spectrophotometer, and generating a standard curve. The report includes the methodology, results presented in tables and graphs, and a discussion of the assay's principles, including the interactions between Coomassie dye and proteins. The discussion also highlights the assay's advantages, limitations, and factors affecting its accuracy, such as the influence of amino acid composition and the need for standardization. The report concludes with an analysis of the results, emphasizing the importance of accurate pipetting and the use of a reagent blank. The document provides a comprehensive overview of the Bradford Protein Assay, making it a valuable resource for students studying biology or related fields.

Biological Sciences 1
BIOLOGICAL SCIENCES
By Name
Course
Instructor
Institution
Location
Date
BIOLOGICAL SCIENCES
By Name
Course
Instructor
Institution
Location
Date
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Biological Sciences 2
Abstract
The Bradford Protein Assay is a spectroscopic procedure of analysis that helps in the
determination of the concentration of proteins in a sample solution. The experiment aims at
measuring the concentration of the unknown protein solutions and after that drawing the standard
curve by plotting the 620 nm against a reagent blank. The standards were prepared through the
addition of various volumes of BSA stock solution. The absorbances of the unknown samples of
protein were determined by the use of a spectrophotometer. A standard curve for the experiment
was achieved by plotting 620 nm results the obtained BSA concentration. Linear regression was
used in the solution of concentrations of the unknown proteins. Different values were obtained
for the various trials that were made, and the average for each trial was calculated.
Introduction
A quicker, simpler and safer procedure used in finding the protein concentration in an aqueous
solution was developed in the 1970s by Marion Bradford, a biochemist. The procedure has been
found to be very efficient thus has become a preference displacing the other methods including
Biuret and Folin, which were originally used in the measurement of soluble proteins (Freshney,
2011, p.159). Because of the disturbance created by the detergents that have been used in the
solubilisation process of the membrane-bound proteins, this procedure is not suitable in the form
described in this experiment.
The original publication of the Bradford Reagent protocol was done in an article Analytical
Biochemistry. The protocol underwent refined modification that made it more useful especially
for membrane-bound proteins. The modifications were published in the same journal article in
the preceding years. The Bradford protein assay is a very convenient method of determining the
Abstract
The Bradford Protein Assay is a spectroscopic procedure of analysis that helps in the
determination of the concentration of proteins in a sample solution. The experiment aims at
measuring the concentration of the unknown protein solutions and after that drawing the standard
curve by plotting the 620 nm against a reagent blank. The standards were prepared through the
addition of various volumes of BSA stock solution. The absorbances of the unknown samples of
protein were determined by the use of a spectrophotometer. A standard curve for the experiment
was achieved by plotting 620 nm results the obtained BSA concentration. Linear regression was
used in the solution of concentrations of the unknown proteins. Different values were obtained
for the various trials that were made, and the average for each trial was calculated.
Introduction
A quicker, simpler and safer procedure used in finding the protein concentration in an aqueous
solution was developed in the 1970s by Marion Bradford, a biochemist. The procedure has been
found to be very efficient thus has become a preference displacing the other methods including
Biuret and Folin, which were originally used in the measurement of soluble proteins (Freshney,
2011, p.159). Because of the disturbance created by the detergents that have been used in the
solubilisation process of the membrane-bound proteins, this procedure is not suitable in the form
described in this experiment.
The original publication of the Bradford Reagent protocol was done in an article Analytical
Biochemistry. The protocol underwent refined modification that made it more useful especially
for membrane-bound proteins. The modifications were published in the same journal article in
the preceding years. The Bradford protein assay is a very convenient method of determining the

Biological Sciences 3
concentration of proteins (David W. Burden, 2012, p.212). It supplies the dye reagent used in the
estimation of the protein concentration at 1x concentrations well as two protein assay standards
at prediluted concentrations of seven.
The seven prediluted standards are efficiently packaged in screwcap vials of volume 2ml. The
packaging reduces and eliminates any form of wasteful sharp ampoules. The packaging also
ensures the protein is stable over the product's shelf life. Bradford Protein Assay measures the
concentration of proteins in a food sample by adding Coomassie dye to the food sample under
acidic conditions. A colour change from brown to blue is noted when proteins in the food sample
bind with the Coomassie dye. The concentration or level of the blue colour can be determined
using spectrophotometer hence determining the protein concentration in the food sample
(Copeland, 2013, p.251).
Among the advantage of Bradford Protein Assay are the few steps involved, no need of heating
as well as its ability to provide a calorimetric response of greater stability. The response of
Bradford Protein Assay method is affected by non-protein sources. It tends to be more nonlinear
towards the greatest concentration of protein range (Freshney, 2011, p.266). The response of this
method is also affected by proteins changes with a change in the composition of the protein
being tested. Due to these limitations, there is need to standardize the solutions of proteins to
achieve results that are more accurate.
The amount of light absorbed by a sample is measured by the use of a spectrophotometer. This
instrument concentrates a beam through a sample thereby taking measurements of the light
intensity that reaches the detector (Blankenship, 2012, p.118). The beam of light is composed of
a stream of photons, which are absorbed by an analyte molecule when they come across each
concentration of proteins (David W. Burden, 2012, p.212). It supplies the dye reagent used in the
estimation of the protein concentration at 1x concentrations well as two protein assay standards
at prediluted concentrations of seven.
The seven prediluted standards are efficiently packaged in screwcap vials of volume 2ml. The
packaging reduces and eliminates any form of wasteful sharp ampoules. The packaging also
ensures the protein is stable over the product's shelf life. Bradford Protein Assay measures the
concentration of proteins in a food sample by adding Coomassie dye to the food sample under
acidic conditions. A colour change from brown to blue is noted when proteins in the food sample
bind with the Coomassie dye. The concentration or level of the blue colour can be determined
using spectrophotometer hence determining the protein concentration in the food sample
(Copeland, 2013, p.251).
Among the advantage of Bradford Protein Assay are the few steps involved, no need of heating
as well as its ability to provide a calorimetric response of greater stability. The response of
Bradford Protein Assay method is affected by non-protein sources. It tends to be more nonlinear
towards the greatest concentration of protein range (Freshney, 2011, p.266). The response of this
method is also affected by proteins changes with a change in the composition of the protein
being tested. Due to these limitations, there is need to standardize the solutions of proteins to
achieve results that are more accurate.
The amount of light absorbed by a sample is measured by the use of a spectrophotometer. This
instrument concentrates a beam through a sample thereby taking measurements of the light
intensity that reaches the detector (Blankenship, 2012, p.118). The beam of light is composed of
a stream of photons, which are absorbed by an analyte molecule when they come across each
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Biological Sciences 4
other. The intensity of the light beam is reduced as more photons are absorbed by the analyte
material during encounters.
The objective of this experiment is to find the concentration of the unknown solution of protein
and draw the standard curve by plotting the 620 nm against a reagent blank.
Methodology
The procedure used in the determination of the presence of protein using Bradford Protein Assay
for the case of this particular experiment was slightly altered with some of the steps skipped, and
others changed to suit the nature of the experiment. The method used was as shown below
Prepare 0.02% stock solution of DCPIP in the tris buffer to a final volume of 50cm3.
Using distilled water as a diluent, prepare a series of dilutions of the stock. Calculate the
final percentage of DCPIP and record the value in the table
Set the spectrophotometer at a wavelength of 620 nm
Zero the machine using a blank control, which has no DCPIP
Measure the absorbance of each dilution and record the values in a table
Generate a curve for the results
Results
Table 1 shows the absorption levels for the two attempts of the experiment and the mean
absorption of DCIP at different percentage concentrations when diluted.
Table 1
Dilution % DCPIP Absorption Absorption Mean
other. The intensity of the light beam is reduced as more photons are absorbed by the analyte
material during encounters.
The objective of this experiment is to find the concentration of the unknown solution of protein
and draw the standard curve by plotting the 620 nm against a reagent blank.
Methodology
The procedure used in the determination of the presence of protein using Bradford Protein Assay
for the case of this particular experiment was slightly altered with some of the steps skipped, and
others changed to suit the nature of the experiment. The method used was as shown below
Prepare 0.02% stock solution of DCPIP in the tris buffer to a final volume of 50cm3.
Using distilled water as a diluent, prepare a series of dilutions of the stock. Calculate the
final percentage of DCPIP and record the value in the table
Set the spectrophotometer at a wavelength of 620 nm
Zero the machine using a blank control, which has no DCPIP
Measure the absorbance of each dilution and record the values in a table
Generate a curve for the results
Results
Table 1 shows the absorption levels for the two attempts of the experiment and the mean
absorption of DCIP at different percentage concentrations when diluted.
Table 1
Dilution % DCPIP Absorption Absorption Mean
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
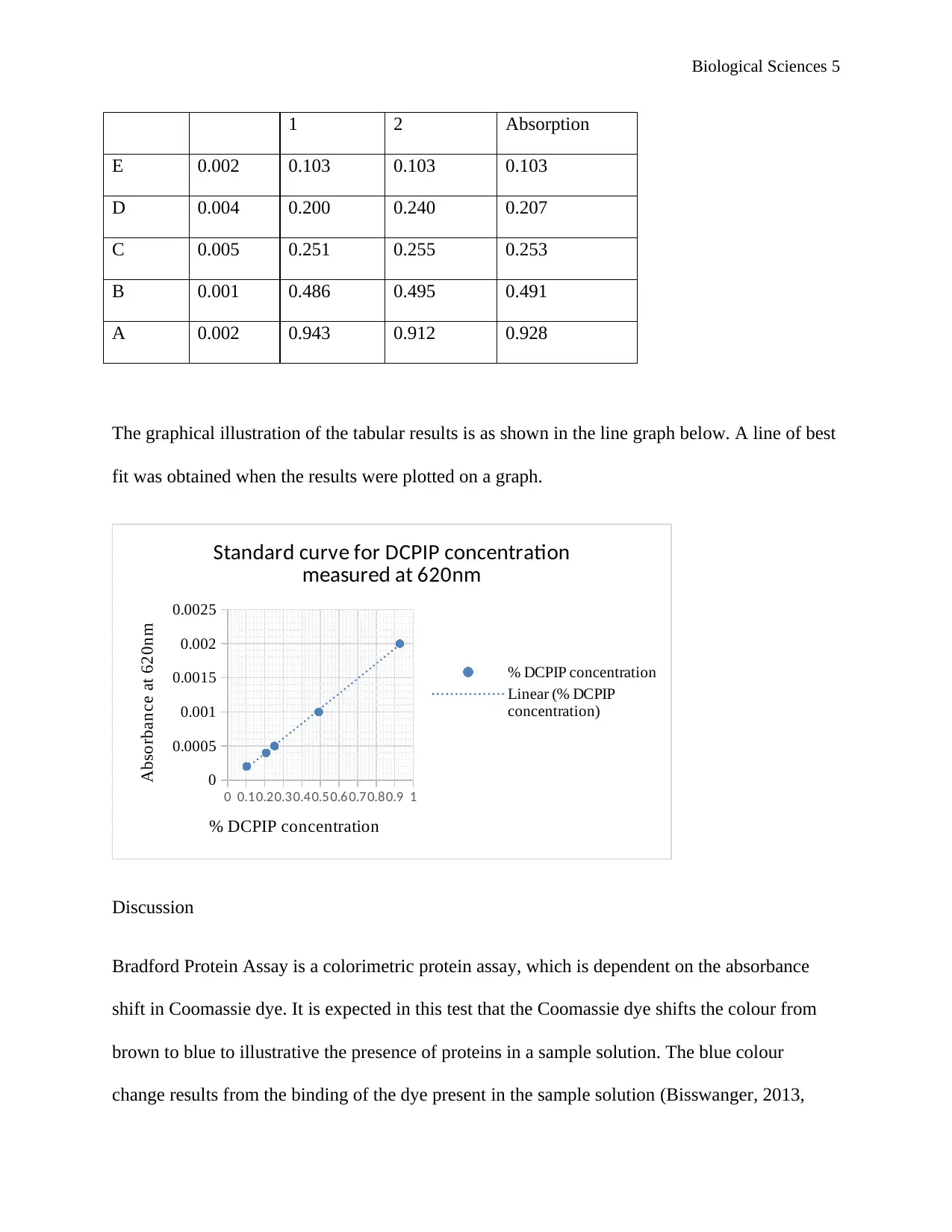
Biological Sciences 5
1 2 Absorption
E 0.002 0.103 0.103 0.103
D 0.004 0.200 0.240 0.207
C 0.005 0.251 0.255 0.253
B 0.001 0.486 0.495 0.491
A 0.002 0.943 0.912 0.928
The graphical illustration of the tabular results is as shown in the line graph below. A line of best
fit was obtained when the results were plotted on a graph.
0 0.10.20.30.40.50.60.70.80.9 1
0
0.0005
0.001
0.0015
0.002
0.0025
Standard curve for DCPIP concentration
measured at 620nm
% DCPIP concentration
Linear (% DCPIP
concentration)
% DCPIP concentration
Absorbance at 620nm
Discussion
Bradford Protein Assay is a colorimetric protein assay, which is dependent on the absorbance
shift in Coomassie dye. It is expected in this test that the Coomassie dye shifts the colour from
brown to blue to illustrative the presence of proteins in a sample solution. The blue colour
change results from the binding of the dye present in the sample solution (Bisswanger, 2013,
1 2 Absorption
E 0.002 0.103 0.103 0.103
D 0.004 0.200 0.240 0.207
C 0.005 0.251 0.255 0.253
B 0.001 0.486 0.495 0.491
A 0.002 0.943 0.912 0.928
The graphical illustration of the tabular results is as shown in the line graph below. A line of best
fit was obtained when the results were plotted on a graph.
0 0.10.20.30.40.50.60.70.80.9 1
0
0.0005
0.001
0.0015
0.002
0.0025
Standard curve for DCPIP concentration
measured at 620nm
% DCPIP concentration
Linear (% DCPIP
concentration)
% DCPIP concentration
Absorbance at 620nm
Discussion
Bradford Protein Assay is a colorimetric protein assay, which is dependent on the absorbance
shift in Coomassie dye. It is expected in this test that the Coomassie dye shifts the colour from
brown to blue to illustrative the presence of proteins in a sample solution. The blue colour
change results from the binding of the dye present in the sample solution (Bisswanger, 2013,

Biological Sciences 6
p.122). Two types of interactions of bonds are observed during the formation of this complex
compound. The first interaction is the donation of a free proton by the brown/red Coomassie dye.
The dye donates its proton to the ionizable group that is on the protein and such a donation
results into a disruption of the native state of the protein thereby leading to the exposure of its
hydrophobic pockets. The exposed pockets, which are on the tertiary structure of the protein non-
covalently, bind to the region of the dye that is not polar by Van der Waals forces. In so doing,
the favourable amino group of the protein is brought close to the negative charge of the dye
(Ganapathy-Kanniappan, 2018, p.351). The ionic interaction between the amino group and the
dye is made stronger by ionic interaction between them. Through binding of the protein, the blue
form of Coomassie dye achieves stability hence the quantity of complex available in the solution
is determined by the concentration of the protein using the reading of the absorbance.
Fig.1. Structure of Coomassie Brilliant Blue
The extinction coefficient remains constant for the complex solution of the dye and protein for
over 10 times the range of concentration (Becker, 2012, p.148). This makes Bradford protein
assay very useful. The concentration-absorbance curve can be changed depending on
hydrophobic or arginine amino acids percentage in each of the proteins. This is because Bradford
protein assay takes measures of the number of residues of arginine or hydrophobic amino acids.
This leads to the need of a standard having a protein having protein proximity to the measured
protein in the composition. An example of such standards is Bovine Serum Albumin (BSA). The
p.122). Two types of interactions of bonds are observed during the formation of this complex
compound. The first interaction is the donation of a free proton by the brown/red Coomassie dye.
The dye donates its proton to the ionizable group that is on the protein and such a donation
results into a disruption of the native state of the protein thereby leading to the exposure of its
hydrophobic pockets. The exposed pockets, which are on the tertiary structure of the protein non-
covalently, bind to the region of the dye that is not polar by Van der Waals forces. In so doing,
the favourable amino group of the protein is brought close to the negative charge of the dye
(Ganapathy-Kanniappan, 2018, p.351). The ionic interaction between the amino group and the
dye is made stronger by ionic interaction between them. Through binding of the protein, the blue
form of Coomassie dye achieves stability hence the quantity of complex available in the solution
is determined by the concentration of the protein using the reading of the absorbance.
Fig.1. Structure of Coomassie Brilliant Blue
The extinction coefficient remains constant for the complex solution of the dye and protein for
over 10 times the range of concentration (Becker, 2012, p.148). This makes Bradford protein
assay very useful. The concentration-absorbance curve can be changed depending on
hydrophobic or arginine amino acids percentage in each of the proteins. This is because Bradford
protein assay takes measures of the number of residues of arginine or hydrophobic amino acids.
This leads to the need of a standard having a protein having protein proximity to the measured
protein in the composition. An example of such standards is Bovine Serum Albumin (BSA). The
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Biological Sciences 7
dye reagent is found to react more with residues of arginine, and the rate of reaction is found to
be lowered with residues of tryptophan, lysine, phenylalanine, tyrosine and histidine. The
accuracy of the assay is considerably lower when measuring basic or acidic proteins (Hock,
2015, p.215). The sensitivity of Bradford to BSA is relatively higher than with average proteins
by approximately a factor of two. 1 M NaOH is added to facilitate solubility of the protein
membranes and also reduce the variations in the yield of colour between a protein and another
protein. Immunoglobin is the most preferred standard of protein.
Any error in the experiment can be a cause of alarm and may interfere with the whole
experiment and its results. It is for this reason that care is taken such that the prediluted standards
are packaged conveniently to eliminate any wasteful and sharp ampoules as well as keep the
shelf life stable (Bisen, 2014, p.188). Care should be taken when pipetting to avoid such errors as
having inadequate or excess of the reagents or the dye. It is also important to have the
spectrophotometer zeroed using the regent blank as it may as well be a source of error in the
experiment. Another precaution is performing not less than two reagents blank or instead use a
water buffer.
Conclusion
The absorbance values that were duplicated in the experiment were close, and the graph turned
out to have a line of best fit. The only significantly different in the values of absorbance was
observed at 0.002% of DCPIP. From the obtained results, it can be understood that the values
have a major increase from Dilution B to Dilution A. this is evidently illustrated on the graph
from the huge shift between 0.001% DCPIP concentration to 0.002% DCPIP concentration.
dye reagent is found to react more with residues of arginine, and the rate of reaction is found to
be lowered with residues of tryptophan, lysine, phenylalanine, tyrosine and histidine. The
accuracy of the assay is considerably lower when measuring basic or acidic proteins (Hock,
2015, p.215). The sensitivity of Bradford to BSA is relatively higher than with average proteins
by approximately a factor of two. 1 M NaOH is added to facilitate solubility of the protein
membranes and also reduce the variations in the yield of colour between a protein and another
protein. Immunoglobin is the most preferred standard of protein.
Any error in the experiment can be a cause of alarm and may interfere with the whole
experiment and its results. It is for this reason that care is taken such that the prediluted standards
are packaged conveniently to eliminate any wasteful and sharp ampoules as well as keep the
shelf life stable (Bisen, 2014, p.188). Care should be taken when pipetting to avoid such errors as
having inadequate or excess of the reagents or the dye. It is also important to have the
spectrophotometer zeroed using the regent blank as it may as well be a source of error in the
experiment. Another precaution is performing not less than two reagents blank or instead use a
water buffer.
Conclusion
The absorbance values that were duplicated in the experiment were close, and the graph turned
out to have a line of best fit. The only significantly different in the values of absorbance was
observed at 0.002% of DCPIP. From the obtained results, it can be understood that the values
have a major increase from Dilution B to Dilution A. this is evidently illustrated on the graph
from the huge shift between 0.001% DCPIP concentration to 0.002% DCPIP concentration.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Biological Sciences 8
References
Becker, J.M., 2012. Biotechnology: A Laboratory Course. 4th ed. Manchester: Academic Press.
Bisen, P.S., 2014. Laboratory Protocols in Applied Life Sciences. 4th ed. Beijing: CRC Press.
Bisswanger, H., 2013. Practical Enzymology. 5th ed. Oxford: John Wiley & Sons.
Blankenship, L., 2012. Colonization Control of Human Bacterial Enteropathogens in Poultry.
5th ed. Oxford: Academic Press.
Copeland, R.A., 2013. Methods for Protein Analysis: A Practical Guide for Laboratory
Protocols. 6th ed. Kansas: Springer Science & Business Media.
David W. Burden, D.B.W., 2012. Biotechnology Proteins to PCR: A Course in Strategies and
Lab Techniques. 5th ed. London: Springer Science & Business Media.
Donglu Zhang, S.S., 2012. ADME-Enabling Technologies in Drug Design and Development. 3rd
ed. Beijing: John Wiley & Sons.
Freshney, R.I., 2011. Culture of Animal Cells: A Manual of Basic Technique and Specialized
Applications. 6th ed. Salt Lake: John Wiley & Sons.
Ganapathy-Kanniappan, S., 2018. Advances in GAPDH Protein Analysis: A Functional and
Biochemical Approach. 3rd ed. Chicago: Springer.
References
Becker, J.M., 2012. Biotechnology: A Laboratory Course. 4th ed. Manchester: Academic Press.
Bisen, P.S., 2014. Laboratory Protocols in Applied Life Sciences. 4th ed. Beijing: CRC Press.
Bisswanger, H., 2013. Practical Enzymology. 5th ed. Oxford: John Wiley & Sons.
Blankenship, L., 2012. Colonization Control of Human Bacterial Enteropathogens in Poultry.
5th ed. Oxford: Academic Press.
Copeland, R.A., 2013. Methods for Protein Analysis: A Practical Guide for Laboratory
Protocols. 6th ed. Kansas: Springer Science & Business Media.
David W. Burden, D.B.W., 2012. Biotechnology Proteins to PCR: A Course in Strategies and
Lab Techniques. 5th ed. London: Springer Science & Business Media.
Donglu Zhang, S.S., 2012. ADME-Enabling Technologies in Drug Design and Development. 3rd
ed. Beijing: John Wiley & Sons.
Freshney, R.I., 2011. Culture of Animal Cells: A Manual of Basic Technique and Specialized
Applications. 6th ed. Salt Lake: John Wiley & Sons.
Ganapathy-Kanniappan, S., 2018. Advances in GAPDH Protein Analysis: A Functional and
Biochemical Approach. 3rd ed. Chicago: Springer.

Biological Sciences 9
Hock, F.J., 2015. Drug Discovery and Evaluation: Pharmacological Assays. 4th ed. London:
Springer International Publishing.
Ninfa, A.J., 2009. Fundamental Laboratory Approaches for Biochemistry and Biotechnology.
5th ed. London: Wiley.
Rosenberg, I.M., 2013. Protein Analysis and Purification: Benchtop Techniques. 2nd ed. New
York: Springer Science & Business Media.
Stephenson, F.H., 2010. Calculations for Molecular Biology and Biotechnology: A Guide to
Mathematics in the Laboratory. 2nd ed. New York: Academic Press.
Thomas E. Crowley, J.K., 2014. Experiments in the Purification and Characterization of
Enzymes: A Laboratory Manual. 3rd ed. New York: Academic Press.
William S. Adney, J.D.M.J.R.M.K.T.K., 2009. Biotechnology for Fuels and Chemicals: The
Twenty-Ninth Symposium. 4th ed. Cambrige: Springer Science & Business Media.
Hock, F.J., 2015. Drug Discovery and Evaluation: Pharmacological Assays. 4th ed. London:
Springer International Publishing.
Ninfa, A.J., 2009. Fundamental Laboratory Approaches for Biochemistry and Biotechnology.
5th ed. London: Wiley.
Rosenberg, I.M., 2013. Protein Analysis and Purification: Benchtop Techniques. 2nd ed. New
York: Springer Science & Business Media.
Stephenson, F.H., 2010. Calculations for Molecular Biology and Biotechnology: A Guide to
Mathematics in the Laboratory. 2nd ed. New York: Academic Press.
Thomas E. Crowley, J.K., 2014. Experiments in the Purification and Characterization of
Enzymes: A Laboratory Manual. 3rd ed. New York: Academic Press.
William S. Adney, J.D.M.J.R.M.K.T.K., 2009. Biotechnology for Fuels and Chemicals: The
Twenty-Ninth Symposium. 4th ed. Cambrige: Springer Science & Business Media.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 9
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2025 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.




