Comprehensive Review: Chromatin Remodeling Complexes and Disease Links
VerifiedAdded on 2021/04/22
|7
|2408
|161
Report
AI Summary
This report provides a detailed overview of chromatin remodeling complexes within eukaryotic cells. It begins with an introduction to cellular biology, differentiating between eukaryotes and prokaryotes, and explaining the role of chromatin. The report then delves into the importance of chromatin remodeling, outlining the functions of remodelers in DNA repair, replication, and transcription. It categorizes chromatin remodeling complexes into ATP-dependent complexes and histone-modifying complexes (HDACs and HATs), with a primary focus on the ATP-dependent families. The report proceeds to examine the four major families of chromatin remodelers: SWI/SNF, INO80, CHD, and ISWI, detailing their composition, functions, and roles in various cellular processes. Furthermore, it explores the interactions of these complexes with DNA, including their involvement in DNA repair, replication initiation, and the progression of replication forks. Finally, the report discusses the links between chromatin remodeling complexes and human diseases, providing examples such as pancreatic cancer, breast cancer, and Charge syndrome. The conclusion emphasizes the ongoing research and the need for further investigation into the complexities of chromatin remodeling and its implications for human health.

NAME:
DATE:
UNIT:
DATE:
UNIT:
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Abstract
Cellular and microbial biology has made great leaps in the study of cells in living organisms and
microorganisms. Studies focused on eukaryotes have led to major discoveries about chromatin,
remodeling of chromatin and its complexes. As much as chromatin modifying complexes are
characterized into ATP-dependent complexes and both Histone acetyltransferase (HAT) and histone
deacetylase complexes (HDACS), scientists have mainly focused on ATP dependent complexes and its
various families. Nonetheless, there have been major contributions on the category of HDACS and HATS
particularly in the study of cancer cells in humans. This review focuses on eukaryotes and takes a keen in
depth focus on the known chromatin remodeling complexes based on their specific functions, the purpose
in which they engage DNA and the links between known chromatin remodeling complexes and human
diseases.
EUKARYOTIC CHROMATIN REMODELING COMPLEXES.
Introduction
In cellular Biology, microorganisms and living organisms can be classified as eukaryotes or prokaryotes
based on their cellular characteristics. For instance, cells of eukaryotes have organelles and a nucleus
while cells of prokaryotes both. Eukaryotes include but not limited to such microorganisms as simple
fungi, algae and protozoa. Eukaryotes have DNA for their genetic formation which is tightly condensed
around proteins called histones into a complex called chromatin. Chromatin remodeling is its
rearrangement from a tightly compressed state to an accessible state by transcription. (Hargreaves, &
Crabtree, 2011). Chromatin therefore needs to be remodeled for the following reasons:
i.) Remodelers adapted for certain functions help slide, restructure, and remove nucleosomes,
when repairing and recombining DNA.
ii.) Remodelers adapted for certain functions are used for correct nucleosome spacing after DNA
is replicated.
iii.) Remodelers assist chromatin to imbue nucleosome vented gaps.
iv.) Exposes a DNA element momentarily on the nucleosome environment.
v.) Allows the factors of transcription and other proteins that bind DNA to control gene
repression and access DNA.
These remodelers are regulated by the following protein complexes:
a.) ATP- dependent complexes that use ATP hydrolysis energy evolved to modify the association
between histones and DNA. They open chromatin structures to facilitate transcriptional activities.
(Narlikar, Sundaramoorthy, & Owen-Hughes, 2013).
b.) Histone deacetylase (HDAC) as well as Histone acetyltransferase (HAT) complexes that regulate the
genes transcription activities through either ubiquitination, acetylation, phosphorylation or methylation
modifications of enzymes.
Currently, studies suggest four families of chromatin remodelers (Bouazoune, Miranda, Jones, &
Kingston, 2009). These families nonetheless are adapted for distinct uses and contexts biologically
yielded by their distinct associated subunits and distinct domains in their catalytic ATPases. These
subunits are anteceded by a letter to represent their origin: mouse(m), human(h), Arabidopsis(a),
Xenopus(x) and Drosophila(d),
These four chromatin families of remodelers are: CHD, SWI/SNF, INO80, and ISWI families.
Cellular and microbial biology has made great leaps in the study of cells in living organisms and
microorganisms. Studies focused on eukaryotes have led to major discoveries about chromatin,
remodeling of chromatin and its complexes. As much as chromatin modifying complexes are
characterized into ATP-dependent complexes and both Histone acetyltransferase (HAT) and histone
deacetylase complexes (HDACS), scientists have mainly focused on ATP dependent complexes and its
various families. Nonetheless, there have been major contributions on the category of HDACS and HATS
particularly in the study of cancer cells in humans. This review focuses on eukaryotes and takes a keen in
depth focus on the known chromatin remodeling complexes based on their specific functions, the purpose
in which they engage DNA and the links between known chromatin remodeling complexes and human
diseases.
EUKARYOTIC CHROMATIN REMODELING COMPLEXES.
Introduction
In cellular Biology, microorganisms and living organisms can be classified as eukaryotes or prokaryotes
based on their cellular characteristics. For instance, cells of eukaryotes have organelles and a nucleus
while cells of prokaryotes both. Eukaryotes include but not limited to such microorganisms as simple
fungi, algae and protozoa. Eukaryotes have DNA for their genetic formation which is tightly condensed
around proteins called histones into a complex called chromatin. Chromatin remodeling is its
rearrangement from a tightly compressed state to an accessible state by transcription. (Hargreaves, &
Crabtree, 2011). Chromatin therefore needs to be remodeled for the following reasons:
i.) Remodelers adapted for certain functions help slide, restructure, and remove nucleosomes,
when repairing and recombining DNA.
ii.) Remodelers adapted for certain functions are used for correct nucleosome spacing after DNA
is replicated.
iii.) Remodelers assist chromatin to imbue nucleosome vented gaps.
iv.) Exposes a DNA element momentarily on the nucleosome environment.
v.) Allows the factors of transcription and other proteins that bind DNA to control gene
repression and access DNA.
These remodelers are regulated by the following protein complexes:
a.) ATP- dependent complexes that use ATP hydrolysis energy evolved to modify the association
between histones and DNA. They open chromatin structures to facilitate transcriptional activities.
(Narlikar, Sundaramoorthy, & Owen-Hughes, 2013).
b.) Histone deacetylase (HDAC) as well as Histone acetyltransferase (HAT) complexes that regulate the
genes transcription activities through either ubiquitination, acetylation, phosphorylation or methylation
modifications of enzymes.
Currently, studies suggest four families of chromatin remodelers (Bouazoune, Miranda, Jones, &
Kingston, 2009). These families nonetheless are adapted for distinct uses and contexts biologically
yielded by their distinct associated subunits and distinct domains in their catalytic ATPases. These
subunits are anteceded by a letter to represent their origin: mouse(m), human(h), Arabidopsis(a),
Xenopus(x) and Drosophila(d),
These four chromatin families of remodelers are: CHD, SWI/SNF, INO80, and ISWI families.
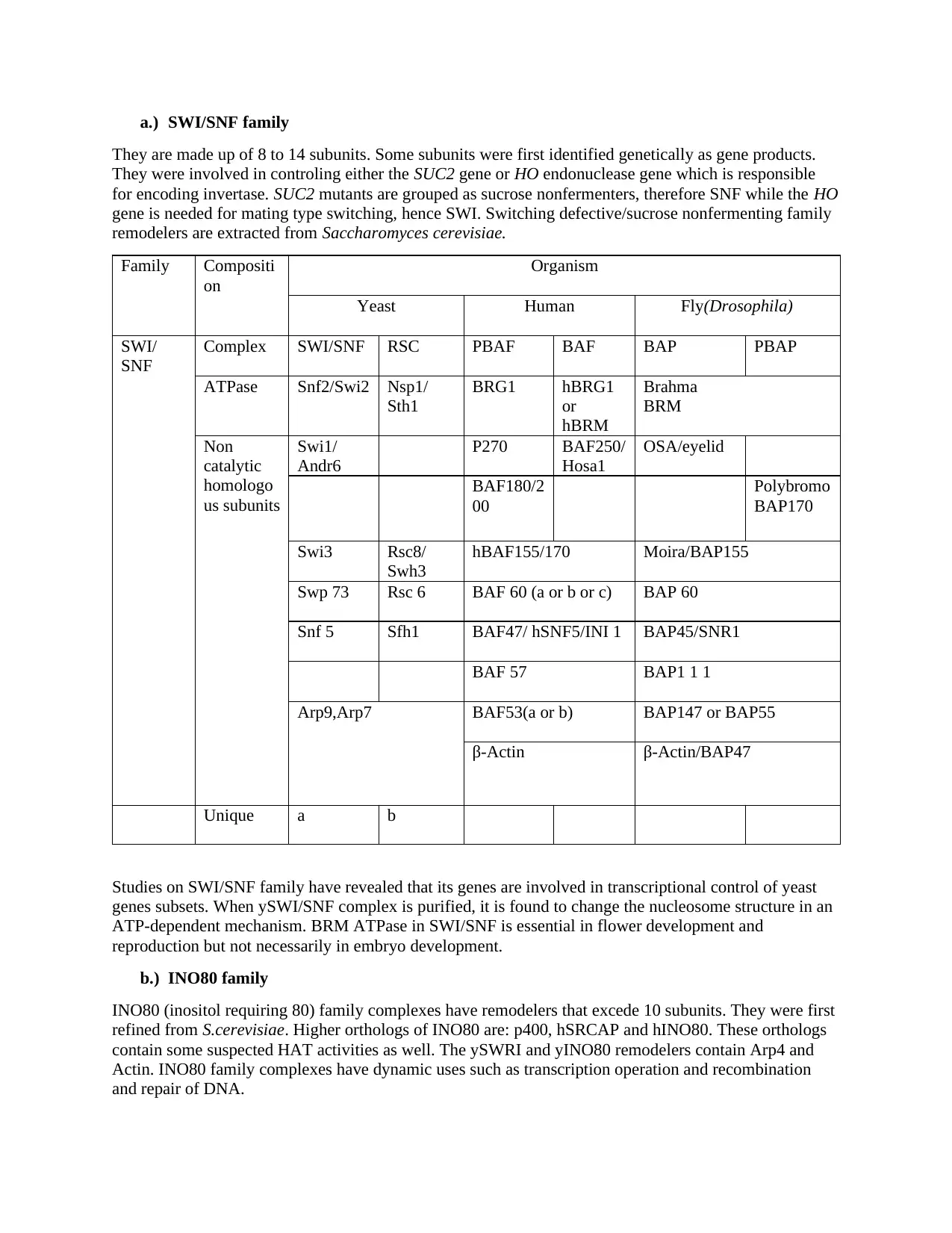
a.) SWI/SNF family
They are made up of 8 to 14 subunits. Some subunits were first identified genetically as gene products.
They were involved in controling either the SUC2 gene or HO endonuclease gene which is responsible
for encoding invertase. SUC2 mutants are grouped as sucrose nonfermenters, therefore SNF while the HO
gene is needed for mating type switching, hence SWI. Switching defective/sucrose nonfermenting family
remodelers are extracted from Saccharomyces cerevisiae.
Family Compositi
on
Organism
Yeast Human Fly(Drosophila)
SWI/
SNF
Complex SWI/SNF RSC PBAF BAF BAP PBAP
ATPase Snf2/Swi2 Nsp1/
Sth1
BRG1 hBRG1
or
hBRM
Brahma
BRM
Non
catalytic
homologo
us subunits
Swi1/
Andr6
P270 BAF250/
Hosa1
OSA/eyelid
BAF180/2
00
Polybromo
BAP170
Swi3 Rsc8/
Swh3
hBAF155/170 Moira/BAP155
Swp 73 Rsc 6 BAF 60 (a or b or c) BAP 60
Snf 5 Sfh1 BAF47/ hSNF5/INI 1 BAP45/SNR1
BAF 57 BAP1 1 1
Arp9,Arp7 BAF53(a or b) BAP147 or BAP55
β-Actin β-Actin/BAP47
Unique a b
Studies on SWI/SNF family have revealed that its genes are involved in transcriptional control of yeast
genes subsets. When ySWI/SNF complex is purified, it is found to change the nucleosome structure in an
ATP-dependent mechanism. BRM ATPase in SWI/SNF is essential in flower development and
reproduction but not necessarily in embryo development.
b.) INO80 family
INO80 (inositol requiring 80) family complexes have remodelers that excede 10 subunits. They were first
refined from S.cerevisiae. Higher orthologs of INO80 are: p400, hSRCAP and hINO80. These orthologs
contain some suspected HAT activities as well. The ySWRI and yINO80 remodelers contain Arp4 and
Actin. INO80 family complexes have dynamic uses such as transcription operation and recombination
and repair of DNA.
They are made up of 8 to 14 subunits. Some subunits were first identified genetically as gene products.
They were involved in controling either the SUC2 gene or HO endonuclease gene which is responsible
for encoding invertase. SUC2 mutants are grouped as sucrose nonfermenters, therefore SNF while the HO
gene is needed for mating type switching, hence SWI. Switching defective/sucrose nonfermenting family
remodelers are extracted from Saccharomyces cerevisiae.
Family Compositi
on
Organism
Yeast Human Fly(Drosophila)
SWI/
SNF
Complex SWI/SNF RSC PBAF BAF BAP PBAP
ATPase Snf2/Swi2 Nsp1/
Sth1
BRG1 hBRG1
or
hBRM
Brahma
BRM
Non
catalytic
homologo
us subunits
Swi1/
Andr6
P270 BAF250/
Hosa1
OSA/eyelid
BAF180/2
00
Polybromo
BAP170
Swi3 Rsc8/
Swh3
hBAF155/170 Moira/BAP155
Swp 73 Rsc 6 BAF 60 (a or b or c) BAP 60
Snf 5 Sfh1 BAF47/ hSNF5/INI 1 BAP45/SNR1
BAF 57 BAP1 1 1
Arp9,Arp7 BAF53(a or b) BAP147 or BAP55
β-Actin β-Actin/BAP47
Unique a b
Studies on SWI/SNF family have revealed that its genes are involved in transcriptional control of yeast
genes subsets. When ySWI/SNF complex is purified, it is found to change the nucleosome structure in an
ATP-dependent mechanism. BRM ATPase in SWI/SNF is essential in flower development and
reproduction but not necessarily in embryo development.
b.) INO80 family
INO80 (inositol requiring 80) family complexes have remodelers that excede 10 subunits. They were first
refined from S.cerevisiae. Higher orthologs of INO80 are: p400, hSRCAP and hINO80. These orthologs
contain some suspected HAT activities as well. The ySWRI and yINO80 remodelers contain Arp4 and
Actin. INO80 family complexes have dynamic uses such as transcription operation and recombination
and repair of DNA.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
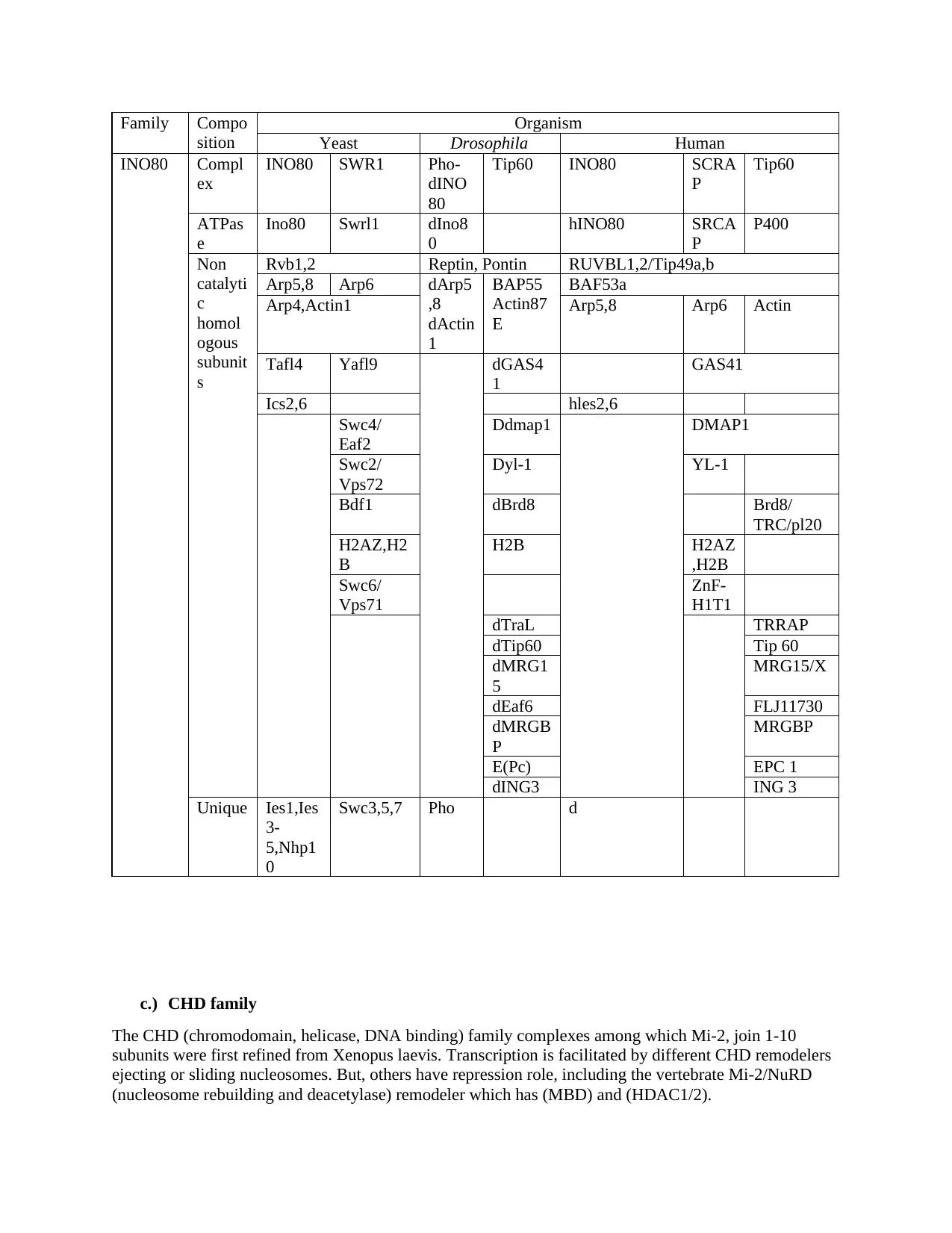
Family Compo
sition
Organism
Yeast Drosophila Human
INO80 Compl
ex
INO80 SWR1 Pho-
dINO
80
Tip60 INO80 SCRA
P
Tip60
ATPas
e
Ino80 Swrl1 dIno8
0
hINO80 SRCA
P
P400
Non
catalyti
c
homol
ogous
subunit
s
Rvb1,2 Reptin, Pontin RUVBL1,2/Tip49a,b
Arp5,8 Arp6 dArp5
,8
dActin
1
BAP55
Actin87
E
BAF53a
Arp4,Actin1 Arp5,8 Arp6 Actin
Tafl4 Yafl9 dGAS4
1
GAS41
Ics2,6 hles2,6
Swc4/
Eaf2
Ddmap1 DMAP1
Swc2/
Vps72
Dyl-1 YL-1
Bdf1 dBrd8 Brd8/
TRC/pl20
H2AZ,H2
B
H2B H2AZ
,H2B
Swc6/
Vps71
ZnF-
H1T1
dTraL TRRAP
dTip60 Tip 60
dMRG1
5
MRG15/X
dEaf6 FLJ11730
dMRGB
P
MRGBP
E(Pc) EPC 1
dING3 ING 3
Unique Ies1,Ies
3-
5,Nhp1
0
Swc3,5,7 Pho d
c.) CHD family
The CHD (chromodomain, helicase, DNA binding) family complexes among which Mi-2, join 1-10
subunits were first refined from Xenopus laevis. Transcription is facilitated by different CHD remodelers
ejecting or sliding nucleosomes. But, others have repression role, including the vertebrate Mi-2/NuRD
(nucleosome rebuilding and deacetylase) remodeler which has (MBD) and (HDAC1/2).
sition
Organism
Yeast Drosophila Human
INO80 Compl
ex
INO80 SWR1 Pho-
dINO
80
Tip60 INO80 SCRA
P
Tip60
ATPas
e
Ino80 Swrl1 dIno8
0
hINO80 SRCA
P
P400
Non
catalyti
c
homol
ogous
subunit
s
Rvb1,2 Reptin, Pontin RUVBL1,2/Tip49a,b
Arp5,8 Arp6 dArp5
,8
dActin
1
BAP55
Actin87
E
BAF53a
Arp4,Actin1 Arp5,8 Arp6 Actin
Tafl4 Yafl9 dGAS4
1
GAS41
Ics2,6 hles2,6
Swc4/
Eaf2
Ddmap1 DMAP1
Swc2/
Vps72
Dyl-1 YL-1
Bdf1 dBrd8 Brd8/
TRC/pl20
H2AZ,H2
B
H2B H2AZ
,H2B
Swc6/
Vps71
ZnF-
H1T1
dTraL TRRAP
dTip60 Tip 60
dMRG1
5
MRG15/X
dEaf6 FLJ11730
dMRGB
P
MRGBP
E(Pc) EPC 1
dING3 ING 3
Unique Ies1,Ies
3-
5,Nhp1
0
Swc3,5,7 Pho d
c.) CHD family
The CHD (chromodomain, helicase, DNA binding) family complexes among which Mi-2, join 1-10
subunits were first refined from Xenopus laevis. Transcription is facilitated by different CHD remodelers
ejecting or sliding nucleosomes. But, others have repression role, including the vertebrate Mi-2/NuRD
(nucleosome rebuilding and deacetylase) remodeler which has (MBD) and (HDAC1/2).
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
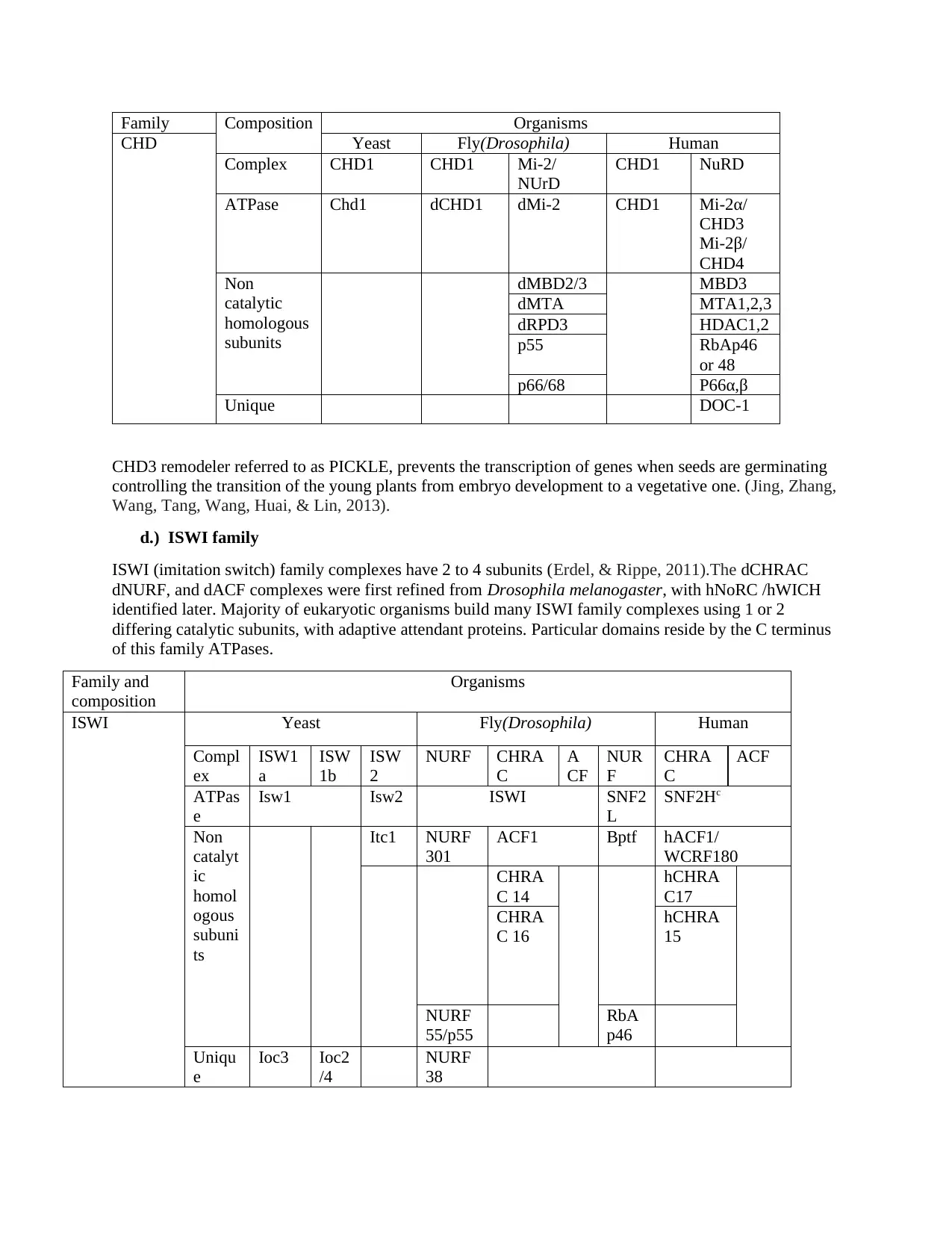
Family Composition Organisms
CHD Yeast Fly(Drosophila) Human
Complex CHD1 CHD1 Mi-2/
NUrD
CHD1 NuRD
ATPase Chd1 dCHD1 dMi-2 CHD1 Mi-2α/
CHD3
Mi-2β/
CHD4
Non
catalytic
homologous
subunits
dMBD2/3 MBD3
dMTA MTA1,2,3
dRPD3 HDAC1,2
p55 RbAp46
or 48
p66/68 P66α,β
Unique DOC-1
CHD3 remodeler referred to as PICKLE, prevents the transcription of genes when seeds are germinating
controlling the transition of the young plants from embryo development to a vegetative one. (Jing, Zhang,
Wang, Tang, Wang, Huai, & Lin, 2013).
d.) ISWI family
ISWI (imitation switch) family complexes have 2 to 4 subunits (Erdel, & Rippe, 2011).The dCHRAC
dNURF, and dACF complexes were first refined from Drosophila melanogaster, with hNoRC /hWICH
identified later. Majority of eukaryotic organisms build many ISWI family complexes using 1 or 2
differing catalytic subunits, with adaptive attendant proteins. Particular domains reside by the C terminus
of this family ATPases.
Family and
composition
Organisms
ISWI Yeast Fly(Drosophila) Human
Compl
ex
ISW1
a
ISW
1b
ISW
2
NURF CHRA
C
A
CF
NUR
F
CHRA
C
ACF
ATPas
e
Isw1 Isw2 ISWI SNF2
L
SNF2Hc
Non
catalyt
ic
homol
ogous
subuni
ts
Itc1 NURF
301
ACF1 Bptf hACF1/
WCRF180
CHRA
C 14
hCHRA
C17
CHRA
C 16
hCHRA
15
NURF
55/p55
RbA
p46
Uniqu
e
Ioc3 Ioc2
/4
NURF
38
CHD Yeast Fly(Drosophila) Human
Complex CHD1 CHD1 Mi-2/
NUrD
CHD1 NuRD
ATPase Chd1 dCHD1 dMi-2 CHD1 Mi-2α/
CHD3
Mi-2β/
CHD4
Non
catalytic
homologous
subunits
dMBD2/3 MBD3
dMTA MTA1,2,3
dRPD3 HDAC1,2
p55 RbAp46
or 48
p66/68 P66α,β
Unique DOC-1
CHD3 remodeler referred to as PICKLE, prevents the transcription of genes when seeds are germinating
controlling the transition of the young plants from embryo development to a vegetative one. (Jing, Zhang,
Wang, Tang, Wang, Huai, & Lin, 2013).
d.) ISWI family
ISWI (imitation switch) family complexes have 2 to 4 subunits (Erdel, & Rippe, 2011).The dCHRAC
dNURF, and dACF complexes were first refined from Drosophila melanogaster, with hNoRC /hWICH
identified later. Majority of eukaryotic organisms build many ISWI family complexes using 1 or 2
differing catalytic subunits, with adaptive attendant proteins. Particular domains reside by the C terminus
of this family ATPases.
Family and
composition
Organisms
ISWI Yeast Fly(Drosophila) Human
Compl
ex
ISW1
a
ISW
1b
ISW
2
NURF CHRA
C
A
CF
NUR
F
CHRA
C
ACF
ATPas
e
Isw1 Isw2 ISWI SNF2
L
SNF2Hc
Non
catalyt
ic
homol
ogous
subuni
ts
Itc1 NURF
301
ACF1 Bptf hACF1/
WCRF180
CHRA
C 14
hCHRA
C17
CHRA
C 16
hCHRA
15
NURF
55/p55
RbA
p46
Uniqu
e
Ioc3 Ioc2
/4
NURF
38

Structure of chromatin complexes
Studies on auxiliary organizations of chromatin remodelers utilizing an electron microscopy with RSC
have demonstrated a major emptiness of nucleosome measurements and the nearness of two compliances
that might be impacted by tail alterations (34-37). Then again, experiments with ySWI/SNF propose
nucleosome binding in a substantial emptiness on the complex surface unveiling component-nucleosome
interaction. The SWI/SNF family edifices, auxiliary and biochemical examinations recommend that the
unit of redesigning is one unpredictable and one nucleosome. Nevertheless, two duplicates of the ISWI
family complex ACF may tie one nucleosome (38, 39), and these two duplicates may physically interface
with each other, raising the likelihood that each ISWI catalyst subunit moves the DNA over the octamer
in the front course, giving a back and forth movement.
DNA engagement with chromatin complexes
Chromatin remodelers not only do they bind DNA but also repair and recombine as well as replication of
DNA. DNA contusions occur frequently debilitate genome integrity, requiring a quick acknowledgment
of locales of harm inside the chromatin. Phosphorylation of serine 139 of H2A.X in vertebrates or serine
129 of H2A in yeast helps enroll DNA repair factors and also chromatin edifices of the INO80 and
SWI/SNF families liable to empower access to DNA ends with a specific end goal to repair enzymes.
(Xiao, Li, Shechter, Ahn , Fabrizio LA, et al., 2009.).) Double-stranded breaks (DSBs) in DNA are
repaired by either of these ways: nonhomologous end joining (NHEJ) or homologous recombination (HR)
The complex ySWI/SNF of the SWI/SNF family fortifies replication beginning in a minichromosome
assesment in yeast (78). Be that as it may, it is yet to be tried in higher orginisms. ISWI remodelers have
broad associations with replication initiation. For example, yISW2 is enhanced at the locales of dynamic
replication and helps with enhancing replication fork movement. (Wilhelm, Barascu, Técher, Debatisse,
& Lopez, 2014).
The remodeler yINO80 of INO80 family effectively adds to typical S phase timing and is very important
for replication fork progression under pressure conditions. yINO80 also enhances fork progression under
typical conditions, relocating alongside PCNA, and helps with keeping up replisome integrity. (Shimada,
Oma, Schleker, Kugou, Ohta, Harata, & Gasser, 2008).
Diseases
The table below gives a description in the links between diseases, syndromes and genetic disorders and
the remodelers involved or suspected to be involved.
Disease Complex involved.
Pancreatic cancer ARID1A and BRG1, SWI/SNF subunits.
Breast cancer Mutations of hSWI/SNF. (Hodges , Kirkland JG,
Crabtree GR, 2016)
Lung cancer SWI/SNF complexes.
Charge syndrome Haploinsufficiency of CHD7
(Berdasco & Esteller, 2013)
ATRX syndrome Mutations in ATRX (SNF2-related ATPase)
(Berdasco & Esteller, 2013)
Williams-Beuren syndrome Increased free hSNF2H
Prostate cancer Chd1(Willmann, Lim, Wetzel, Metzger,
Jandausch, Wilk, & Kirfel,2012)
Conclusion
Studies on auxiliary organizations of chromatin remodelers utilizing an electron microscopy with RSC
have demonstrated a major emptiness of nucleosome measurements and the nearness of two compliances
that might be impacted by tail alterations (34-37). Then again, experiments with ySWI/SNF propose
nucleosome binding in a substantial emptiness on the complex surface unveiling component-nucleosome
interaction. The SWI/SNF family edifices, auxiliary and biochemical examinations recommend that the
unit of redesigning is one unpredictable and one nucleosome. Nevertheless, two duplicates of the ISWI
family complex ACF may tie one nucleosome (38, 39), and these two duplicates may physically interface
with each other, raising the likelihood that each ISWI catalyst subunit moves the DNA over the octamer
in the front course, giving a back and forth movement.
DNA engagement with chromatin complexes
Chromatin remodelers not only do they bind DNA but also repair and recombine as well as replication of
DNA. DNA contusions occur frequently debilitate genome integrity, requiring a quick acknowledgment
of locales of harm inside the chromatin. Phosphorylation of serine 139 of H2A.X in vertebrates or serine
129 of H2A in yeast helps enroll DNA repair factors and also chromatin edifices of the INO80 and
SWI/SNF families liable to empower access to DNA ends with a specific end goal to repair enzymes.
(Xiao, Li, Shechter, Ahn , Fabrizio LA, et al., 2009.).) Double-stranded breaks (DSBs) in DNA are
repaired by either of these ways: nonhomologous end joining (NHEJ) or homologous recombination (HR)
The complex ySWI/SNF of the SWI/SNF family fortifies replication beginning in a minichromosome
assesment in yeast (78). Be that as it may, it is yet to be tried in higher orginisms. ISWI remodelers have
broad associations with replication initiation. For example, yISW2 is enhanced at the locales of dynamic
replication and helps with enhancing replication fork movement. (Wilhelm, Barascu, Técher, Debatisse,
& Lopez, 2014).
The remodeler yINO80 of INO80 family effectively adds to typical S phase timing and is very important
for replication fork progression under pressure conditions. yINO80 also enhances fork progression under
typical conditions, relocating alongside PCNA, and helps with keeping up replisome integrity. (Shimada,
Oma, Schleker, Kugou, Ohta, Harata, & Gasser, 2008).
Diseases
The table below gives a description in the links between diseases, syndromes and genetic disorders and
the remodelers involved or suspected to be involved.
Disease Complex involved.
Pancreatic cancer ARID1A and BRG1, SWI/SNF subunits.
Breast cancer Mutations of hSWI/SNF. (Hodges , Kirkland JG,
Crabtree GR, 2016)
Lung cancer SWI/SNF complexes.
Charge syndrome Haploinsufficiency of CHD7
(Berdasco & Esteller, 2013)
ATRX syndrome Mutations in ATRX (SNF2-related ATPase)
(Berdasco & Esteller, 2013)
Williams-Beuren syndrome Increased free hSNF2H
Prostate cancer Chd1(Willmann, Lim, Wetzel, Metzger,
Jandausch, Wilk, & Kirfel,2012)
Conclusion
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Major discoveries have been made in the field of cellular biology particularly in chromatin complexes
remodelers but a lot of information lies in the balance between certainty and uncertainty. Many of the
already made hypothesis have to be tested and proved scientifically for more fruitful results for major
leaps in research particularly on diseases that undergo complex remodeler mutations.
References
1.) Bouazoune, K., Miranda, T. B., Jones, P. A., & Kingston, R. E. (2009). Analysis of individual
remodeled nucleosomes reveals decreased histone–DNA contacts created by
hSWI/SNF. Nucleic acids research, 37(16), 5279-5294.
2.) Hargreaves, D. C., & Crabtree, G. R. (2011). ATP-dependent chromatin remodeling: genetics,
genomics and mechanisms. Cell research, 21(3), 396.
3.) Narlikar, G. J., Sundaramoorthy, R., & Owen-Hughes, T. (2013). Mechanisms and functions of
ATP-dependent chromatin-remodeling enzymes. Cell, 154(3), 490-503.
4.) Jing, Y., Zhang, D., Wang, X., Tang, W., Wang, W., Huai, J. & Lin, R. (2013). Arabidopsis
chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate
hypocotyl cell elongation. The Plant Cell, 25(1), 242-256.
5.) Erdel, F., & Rippe, K. (2011). Chromatin remodelling in mammalian cells by ISWI‐type
complexes–where, when and why? The FEBS journal, 278(19), 3608-3618.
6.) Xiao, A., Li, H., Shechter, D., Ahn, S. H., Fabrizio, L. A., Erdjument-Bromage, H & Patel, D. J.
(2009). WSTF regulates the H2A. X DNA damage response via a novel tyrosine kinase
activity. Nature, 457(7225), 57.
7.) Wilhelm, T., Barascu, A., Técher, H., Debatisse, M., & Lopez, B. S. (2014). Spontaneous slow
replication fork progression elicits mitosis alterations in homologous recombination-deficient
mammalian cells. Proceedings of the National Academy of Sciences, 111(2), 763-768.
8.) Hodges, C., Kirkland, J. G., & Crabtree, G. R. (2016). The many roles of BAF (mSWI/SNF) and
PBAF complexes in cancer. Cold Spring Harbor perspectives in medicine, 6(8).
9.) Dawson, M. A., & Kouzarides, T. (2012). Cancer epigenetics: from mechanism to
therapy. Cell, 150(1), 12-27.
10.)Zhang, H., Rider, S. D., Henderson, J. T., Fountain, M., Chuang, K., Kandachar, V. & Ogas, J.
(2008). The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. Journal
of Biological Chemistry, 283(33), 22637-22648.
11.)Shimada, K., Oma, Y., Schleker, T., Kugou, K., Ohta, K., Harata, M., & Gasser, S. M. (2008).
Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Current
Biology, 18(8), 566-575.
12.)Willmann, D., Lim, S., Wetzel, S., Metzger, E., Jandausch, A., Wilk, W. & Kirfel, J. (2012).
Impairment of prostate cancer cell growth by a selective and reversible lysine‐specific
demethylase 1 inhibitor. International journal of cancer, 131(11), 2704-2709.
13.)Berdasco, M., & Esteller, M. (2013). Genetic syndromes caused by mutations in epigenetic
genes. Human genetics, 132(4), 359-383
remodelers but a lot of information lies in the balance between certainty and uncertainty. Many of the
already made hypothesis have to be tested and proved scientifically for more fruitful results for major
leaps in research particularly on diseases that undergo complex remodeler mutations.
References
1.) Bouazoune, K., Miranda, T. B., Jones, P. A., & Kingston, R. E. (2009). Analysis of individual
remodeled nucleosomes reveals decreased histone–DNA contacts created by
hSWI/SNF. Nucleic acids research, 37(16), 5279-5294.
2.) Hargreaves, D. C., & Crabtree, G. R. (2011). ATP-dependent chromatin remodeling: genetics,
genomics and mechanisms. Cell research, 21(3), 396.
3.) Narlikar, G. J., Sundaramoorthy, R., & Owen-Hughes, T. (2013). Mechanisms and functions of
ATP-dependent chromatin-remodeling enzymes. Cell, 154(3), 490-503.
4.) Jing, Y., Zhang, D., Wang, X., Tang, W., Wang, W., Huai, J. & Lin, R. (2013). Arabidopsis
chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate
hypocotyl cell elongation. The Plant Cell, 25(1), 242-256.
5.) Erdel, F., & Rippe, K. (2011). Chromatin remodelling in mammalian cells by ISWI‐type
complexes–where, when and why? The FEBS journal, 278(19), 3608-3618.
6.) Xiao, A., Li, H., Shechter, D., Ahn, S. H., Fabrizio, L. A., Erdjument-Bromage, H & Patel, D. J.
(2009). WSTF regulates the H2A. X DNA damage response via a novel tyrosine kinase
activity. Nature, 457(7225), 57.
7.) Wilhelm, T., Barascu, A., Técher, H., Debatisse, M., & Lopez, B. S. (2014). Spontaneous slow
replication fork progression elicits mitosis alterations in homologous recombination-deficient
mammalian cells. Proceedings of the National Academy of Sciences, 111(2), 763-768.
8.) Hodges, C., Kirkland, J. G., & Crabtree, G. R. (2016). The many roles of BAF (mSWI/SNF) and
PBAF complexes in cancer. Cold Spring Harbor perspectives in medicine, 6(8).
9.) Dawson, M. A., & Kouzarides, T. (2012). Cancer epigenetics: from mechanism to
therapy. Cell, 150(1), 12-27.
10.)Zhang, H., Rider, S. D., Henderson, J. T., Fountain, M., Chuang, K., Kandachar, V. & Ogas, J.
(2008). The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. Journal
of Biological Chemistry, 283(33), 22637-22648.
11.)Shimada, K., Oma, Y., Schleker, T., Kugou, K., Ohta, K., Harata, M., & Gasser, S. M. (2008).
Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Current
Biology, 18(8), 566-575.
12.)Willmann, D., Lim, S., Wetzel, S., Metzger, E., Jandausch, A., Wilk, W. & Kirfel, J. (2012).
Impairment of prostate cancer cell growth by a selective and reversible lysine‐specific
demethylase 1 inhibitor. International journal of cancer, 131(11), 2704-2709.
13.)Berdasco, M., & Esteller, M. (2013). Genetic syndromes caused by mutations in epigenetic
genes. Human genetics, 132(4), 359-383
1 out of 7
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.



