CYP3A4 Inhibitor Synthesis, Structure-Activity Relationship Analysis
VerifiedAdded on 2022/09/09
|15
|2196
|16
Homework Assignment
AI Summary
This assignment delves into the study of CYP3A4 inhibitors, focusing on the synthesis of ritonavir-like compounds and their interactions with the CYP3A4 enzyme. The assignment begins with a risk assessment of a synthesis step, considering safety precautions and waste disposal. It then proceeds to analyze 1H NMR spectral data, including peak assignments and interpretations. Furthermore, the assignment explores the binding interactions of the synthesized compounds with the CYP3A4 enzyme, utilizing the Protein Data Bank to identify non-bonding interactions and complete a table. Finally, the assignment examines the relationship between the structure of the inhibitors and their binding affinity, discussing the impact of structural changes on IC50 values and the relevance to the pharmacophore model. The assignment also explores the catalytic cycle of CYP3A4 and compares the inhibitory properties of erythromycin and ritonavir, explaining the differences in their mechanisms of action.
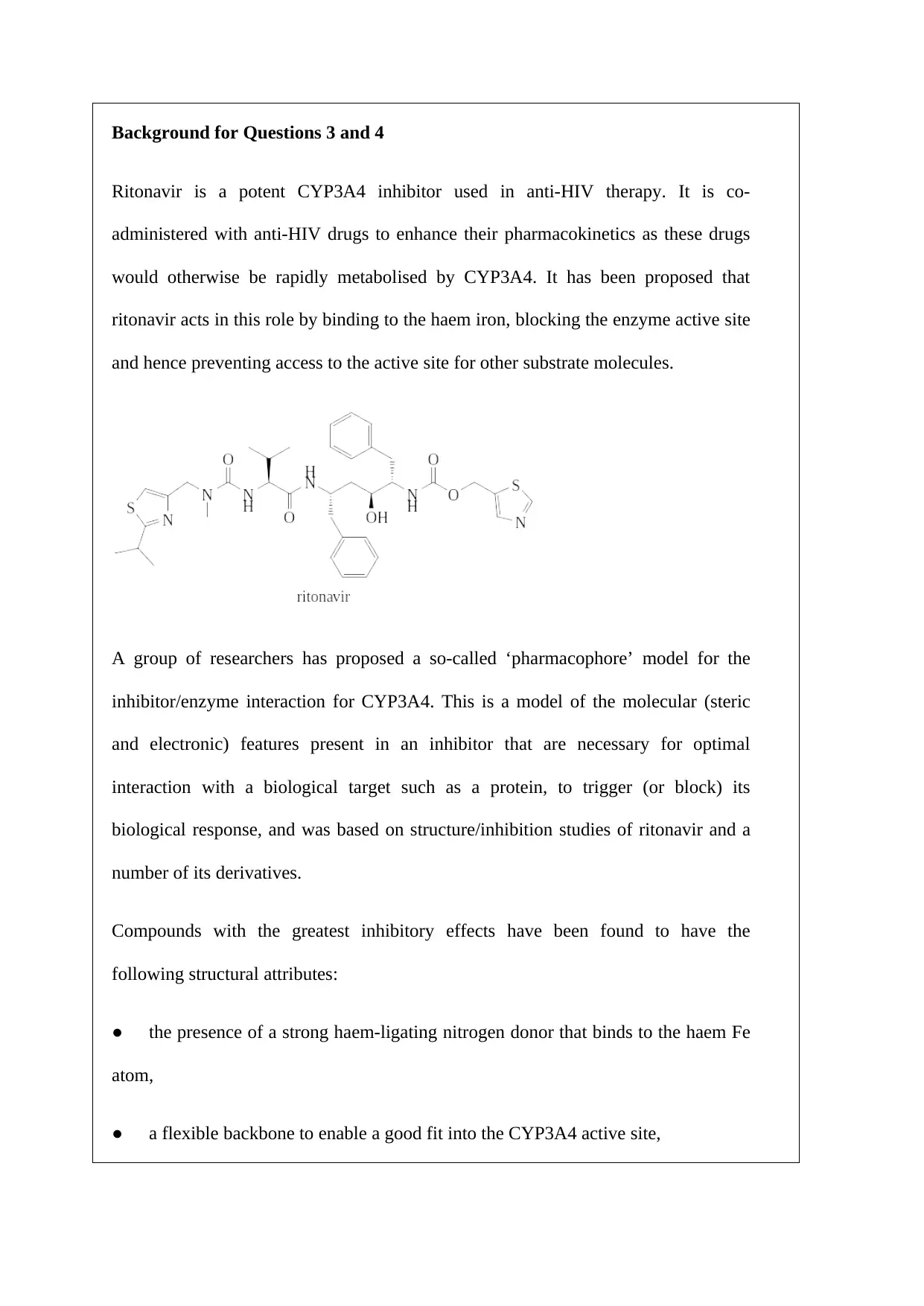
Background for Questions 3 and 4
Ritonavir is a potent CYP3A4 inhibitor used in anti-HIV therapy. It is co-
administered with anti-HIV drugs to enhance their pharmacokinetics as these drugs
would otherwise be rapidly metabolised by CYP3A4. It has been proposed that
ritonavir acts in this role by binding to the haem iron, blocking the enzyme active site
and hence preventing access to the active site for other substrate molecules.
A group of researchers has proposed a so-called ‘pharmacophore’ model for the
inhibitor/enzyme interaction for CYP3A4. This is a model of the molecular (steric
and electronic) features present in an inhibitor that are necessary for optimal
interaction with a biological target such as a protein, to trigger (or block) its
biological response, and was based on structure/inhibition studies of ritonavir and a
number of its derivatives.
Compounds with the greatest inhibitory effects have been found to have the
following structural attributes:
● the presence of a strong haem-ligating nitrogen donor that binds to the haem Fe
atom,
● a flexible backbone to enable a good fit into the CYP3A4 active site,
Ritonavir is a potent CYP3A4 inhibitor used in anti-HIV therapy. It is co-
administered with anti-HIV drugs to enhance their pharmacokinetics as these drugs
would otherwise be rapidly metabolised by CYP3A4. It has been proposed that
ritonavir acts in this role by binding to the haem iron, blocking the enzyme active site
and hence preventing access to the active site for other substrate molecules.
A group of researchers has proposed a so-called ‘pharmacophore’ model for the
inhibitor/enzyme interaction for CYP3A4. This is a model of the molecular (steric
and electronic) features present in an inhibitor that are necessary for optimal
interaction with a biological target such as a protein, to trigger (or block) its
biological response, and was based on structure/inhibition studies of ritonavir and a
number of its derivatives.
Compounds with the greatest inhibitory effects have been found to have the
following structural attributes:
● the presence of a strong haem-ligating nitrogen donor that binds to the haem Fe
atom,
● a flexible backbone to enable a good fit into the CYP3A4 active site,
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

● hydrophobic side groups that can interact with hydrophobic pockets in the active
site, including through the use of π-interactions,
● a hydrogen acceptor/donor capable of hydrogen bonding interactions with
protein residues near the active site.
The researchers synthesised a series of ritonavir-like compounds to evaluate this
model. The general structure of the compounds is shown below.
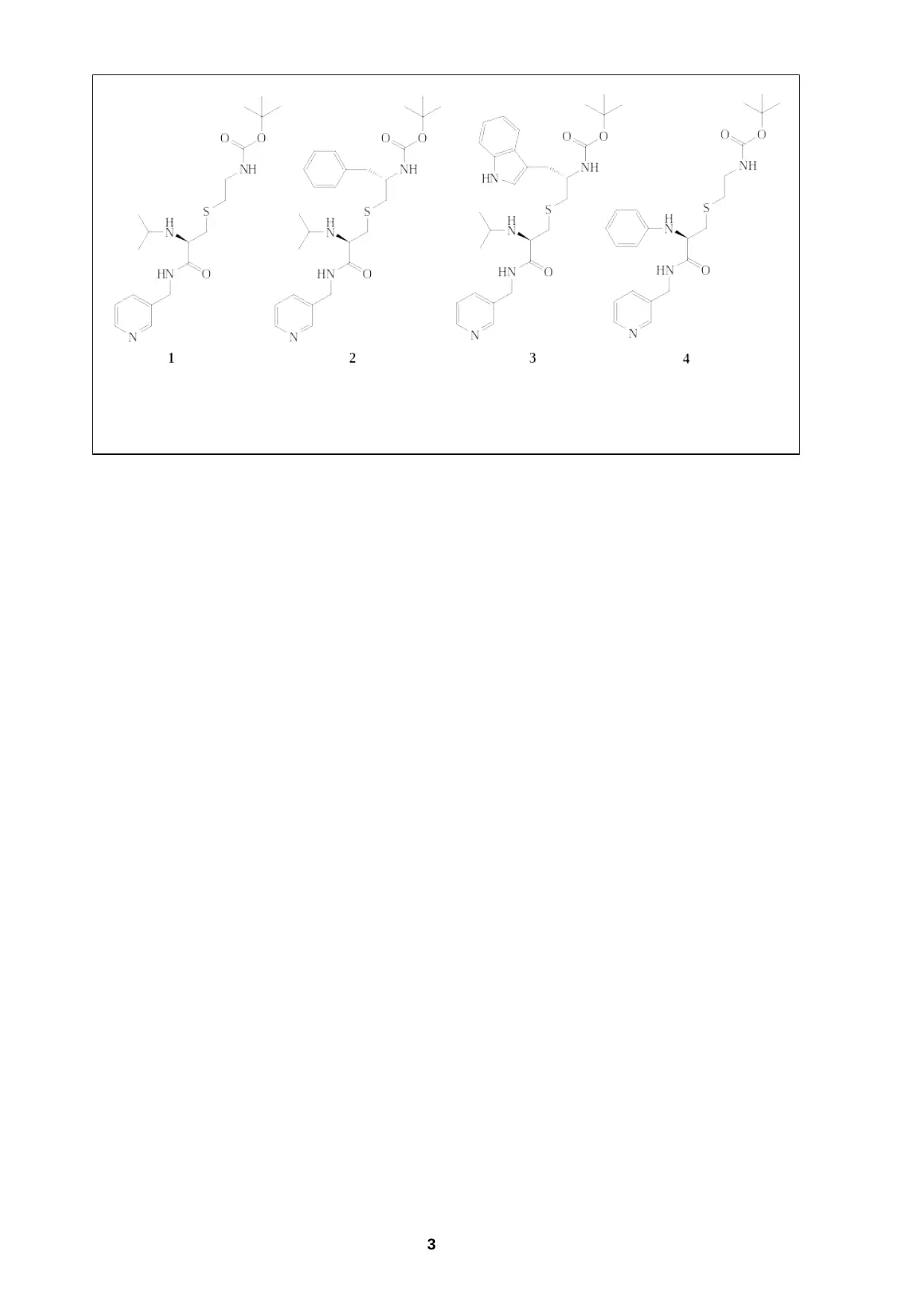
In this study, the researchers made a series of modifications to the groups at the R1
and R2 positions. Four of these structures from this range of compounds are shown
below. The researchers probed how stepwise changes to the structure contributed to
the binding affinity and inhibition properties of the molecules, as well as their effects
on the crystal structure of the CYP3A4 enzyme on binding.
2
site, including through the use of π-interactions,
● a hydrogen acceptor/donor capable of hydrogen bonding interactions with
protein residues near the active site.
The researchers synthesised a series of ritonavir-like compounds to evaluate this
model. The general structure of the compounds is shown below.
In this study, the researchers made a series of modifications to the groups at the R1
and R2 positions. Four of these structures from this range of compounds are shown
below. The researchers probed how stepwise changes to the structure contributed to
the binding affinity and inhibition properties of the molecules, as well as their effects
on the crystal structure of the CYP3A4 enzyme on binding.
2
3
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Question 3
This question carries 29 marks for this assignment and tests learning outcomes KU1,
KU2–KU4, CS1, CS2, KS1, KS3, PS1 and PS2.,
(a) The following scheme shows two steps for the synthesis of a phenyl substituent at the
R1 position in compound 4. The experimental procedure for the first step of the
synthesis involves reactants A and B, and product C as given below.
2-Bromoacrylic acid (7.3 mmol), compound B, was added to a solution of
thioacetanilide (6.6 mmol), compound A, in dry toluene (20 ml) where the mixture
was stirred at 90 °C for 1 h, and then allowed to cool to room temperature. The
intermediate compound C precipitated out of solution and was filtered, washed with
acetone, and recrystallized from MeOH:EtOAc:hexane (1:1:2) to afford the pure
thiazolinium bromide intermediate (compound C).
Prior to completing an experimental procedure such as this it is necessary to carry out
a risk assessment.
Discuss in your own words for the experimental procedure of the first step of the
synthesis, giving your reasons, any safety precautions that you would take when
performing this reaction. You do NOT need to consider the recrystallization of this
product (i.e. the steps following the washing of the filtrate) or the second reaction step
in your answer.
4
This question carries 29 marks for this assignment and tests learning outcomes KU1,
KU2–KU4, CS1, CS2, KS1, KS3, PS1 and PS2.,
(a) The following scheme shows two steps for the synthesis of a phenyl substituent at the
R1 position in compound 4. The experimental procedure for the first step of the
synthesis involves reactants A and B, and product C as given below.
2-Bromoacrylic acid (7.3 mmol), compound B, was added to a solution of
thioacetanilide (6.6 mmol), compound A, in dry toluene (20 ml) where the mixture
was stirred at 90 °C for 1 h, and then allowed to cool to room temperature. The
intermediate compound C precipitated out of solution and was filtered, washed with
acetone, and recrystallized from MeOH:EtOAc:hexane (1:1:2) to afford the pure
thiazolinium bromide intermediate (compound C).
Prior to completing an experimental procedure such as this it is necessary to carry out
a risk assessment.
Discuss in your own words for the experimental procedure of the first step of the
synthesis, giving your reasons, any safety precautions that you would take when
performing this reaction. You do NOT need to consider the recrystallization of this
product (i.e. the steps following the washing of the filtrate) or the second reaction step
in your answer.
4
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Compound C is a novel compound generated in the synthesis and so no Safety data
sheet is available for this compound. In such cases it is standard practice that you treat
this compound (and indeed any new compound or by-product) as toxic unless serious
hazards can be anticipated which is not the case in this instance.
For the risk assessment you should consider:
where you would perform the synthesis of compound C,
what personal protection should be worn during the procedure,
and how you would dispose of any waste.
Material Safety Data Sheets for thioacetanilide (compound A), 2-bromoacrylic acid
(compound B), toluene and acetone are supplied in the assessment folder on the website
for your use. Your answer should be approximately 150 - 200 words.
(7 marks)
5
sheet is available for this compound. In such cases it is standard practice that you treat
this compound (and indeed any new compound or by-product) as toxic unless serious
hazards can be anticipated which is not the case in this instance.
For the risk assessment you should consider:
where you would perform the synthesis of compound C,
what personal protection should be worn during the procedure,
and how you would dispose of any waste.
Material Safety Data Sheets for thioacetanilide (compound A), 2-bromoacrylic acid
(compound B), toluene and acetone are supplied in the assessment folder on the website
for your use. Your answer should be approximately 150 - 200 words.
(7 marks)
5

(b) The 1H NMR spectral data of compound 2 is given in Table 1.
Table 1 400 MHz 1H NMR spectral data for compound 2
δ/ppm* Proton
number
1H NMR
(400 MHz, CD3OD
with a few drops of
D2O)
8.75 (1H, s)
7.85 (1H, d, J = 5.1 Hz)
7.63 (1H, d, J = 7.9 Hz)
7.41–7.38 (1H, dd, J = 5.1, 7.9 Hz)
7.21- 7.18 (2H, dd, J = 7.5, 7.7 Hz)
7.10 (1H, d, J = 7.5 Hz)
7.03 (2H, d, J = 7.7 Hz)
3.48 (2H, d)
3.30 (3H, m)
3.24 (1H, m)
3.04 (1H, m)
4
18
12
5 and 7
9
6
Table 1 400 MHz 1H NMR spectral data for compound 2
δ/ppm* Proton
number
1H NMR
(400 MHz, CD3OD
with a few drops of
D2O)
8.75 (1H, s)
7.85 (1H, d, J = 5.1 Hz)
7.63 (1H, d, J = 7.9 Hz)
7.41–7.38 (1H, dd, J = 5.1, 7.9 Hz)
7.21- 7.18 (2H, dd, J = 7.5, 7.7 Hz)
7.10 (1H, d, J = 7.5 Hz)
7.03 (2H, d, J = 7.7 Hz)
3.48 (2H, d)
3.30 (3H, m)
3.24 (1H, m)
3.04 (1H, m)
4
18
12
5 and 7
9
6
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

2.74 (2H, d)
2.68 (2H, d)
1.63 (9H, s)
1.04 (6H, d)
13
15
22, 23, & 24
*s = singlet, d = doublet, dd = doublet of doublets, m = unresolved multiplet.
The resolution of the spectrum is such that no more than 3J coupling data is observed.
(i) Sketch the splitting pattern for the signal at 7.41-7.38 ppm to show how this can
be assigned to the proton at position 3 (H3). Label your sketch with the
appropriate coupling constants, suggesting which value is associated with
coupling with which proton.
(5 marks)
(ii) When the spectrum was run in CDCl3 without D2O, three additional broad peaks
were observed between 3 and 7 ppm. Suggest the origin of these signals and
briefly explain (one or two sentences) why these were not observed in the
spectrum given in Table 1.
(3 marks)
(iii) Use the S315 data tables and the information in Section 5.3 Interpreting and
predicting NMR spectra to complete Table 1, assigning the remaining peaks in
the spectrum for compound 2. Include a brief explanation (one or two sentences
for each signal assigned) in your answer, considering the integral, multiplicity
and value of the coupling constant(s) where appropriate.
(14 marks)
7
2.68 (2H, d)
1.63 (9H, s)
1.04 (6H, d)
13
15
22, 23, & 24
*s = singlet, d = doublet, dd = doublet of doublets, m = unresolved multiplet.
The resolution of the spectrum is such that no more than 3J coupling data is observed.
(i) Sketch the splitting pattern for the signal at 7.41-7.38 ppm to show how this can
be assigned to the proton at position 3 (H3). Label your sketch with the
appropriate coupling constants, suggesting which value is associated with
coupling with which proton.
(5 marks)
(ii) When the spectrum was run in CDCl3 without D2O, three additional broad peaks
were observed between 3 and 7 ppm. Suggest the origin of these signals and
briefly explain (one or two sentences) why these were not observed in the
spectrum given in Table 1.
(3 marks)
(iii) Use the S315 data tables and the information in Section 5.3 Interpreting and
predicting NMR spectra to complete Table 1, assigning the remaining peaks in
the spectrum for compound 2. Include a brief explanation (one or two sentences
for each signal assigned) in your answer, considering the integral, multiplicity
and value of the coupling constant(s) where appropriate.
(14 marks)
7
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Question 4
This question carries 18 marks for this assignment and tests learning outcomes KU1,
KU2, KU4, CS1, CS2, KS1, KS2 and KS5.
(a) Compounds 1, 2 and 3 are each bound to the haem iron of CYP3A4 via their pyridinic
nitrogen, however, each compound has a different group at the R2 position. The crystal
structures for compounds 1, 2 and 3 bound to the CYP3A4 enzyme have been
deposited in the RCSB Protein Data Bank. Explore the non-bonding interactions
between the substituent group (see dashed boxes on the model compound below) of
the respective compounds 1, 2 and 3, and the CYP3A4 enzyme using the Ligand
Viewer tool for each compound (see instructions below Table 2).
8
This question carries 18 marks for this assignment and tests learning outcomes KU1,
KU2, KU4, CS1, CS2, KS1, KS2 and KS5.
(a) Compounds 1, 2 and 3 are each bound to the haem iron of CYP3A4 via their pyridinic
nitrogen, however, each compound has a different group at the R2 position. The crystal
structures for compounds 1, 2 and 3 bound to the CYP3A4 enzyme have been
deposited in the RCSB Protein Data Bank. Explore the non-bonding interactions
between the substituent group (see dashed boxes on the model compound below) of
the respective compounds 1, 2 and 3, and the CYP3A4 enzyme using the Ligand
Viewer tool for each compound (see instructions below Table 2).
8
The amino acid residues and the haem group on the enzyme can be involved in
hydrogen bonding, hydrophobic contacts or π-interactions (which can either be π−π
interactions, π−cation or π−lone pair interactions) with the various groups of the
inhibitor compounds 1, 2 and 3 - these are listed for compound 1 and 3 in Table 2.
Complete this table for compound 2. (Note The crystal structure for compound 3 is not
very well resolved, i.e. it is disordered, hence the options shown in selection for the
respective interactions with R1 (see Table 2 below)).
Table 2 PDB and ligand ID for compounds 1–3
Compoun
d
PDB code (ligan
d ID)
Hydrophobic
contacts
H-bonds π-π stacking or
π−cation or π-lone pair
interactions
1 6BCZ (D7Y) THR309 -
Pyridine
ALA305 -
Pyridine
ILE120 -R1
Two contacts
with PHE108 –
R1
PHE 213-
PHE241,
PHE213-
PHE304,
ILE301-
PHE304,
SER119 –
Carbonyl
6, ILE301-
ALA305,
ALA305-
THR309,
ALA370-
ARG372
PHE108 - R1 (π- cation)
9
hydrogen bonding, hydrophobic contacts or π-interactions (which can either be π−π
interactions, π−cation or π−lone pair interactions) with the various groups of the
inhibitor compounds 1, 2 and 3 - these are listed for compound 1 and 3 in Table 2.
Complete this table for compound 2. (Note The crystal structure for compound 3 is not
very well resolved, i.e. it is disordered, hence the options shown in selection for the
respective interactions with R1 (see Table 2 below)).
Table 2 PDB and ligand ID for compounds 1–3
Compoun
d
PDB code (ligan
d ID)
Hydrophobic
contacts
H-bonds π-π stacking or
π−cation or π-lone pair
interactions
1 6BCZ (D7Y) THR309 -
Pyridine
ALA305 -
Pyridine
ILE120 -R1
Two contacts
with PHE108 –
R1
PHE 213-
PHE241,
PHE213-
PHE304,
ILE301-
PHE304,
SER119 –
Carbonyl
6, ILE301-
ALA305,
ALA305-
THR309,
ALA370-
ARG372
PHE108 - R1 (π- cation)
9
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
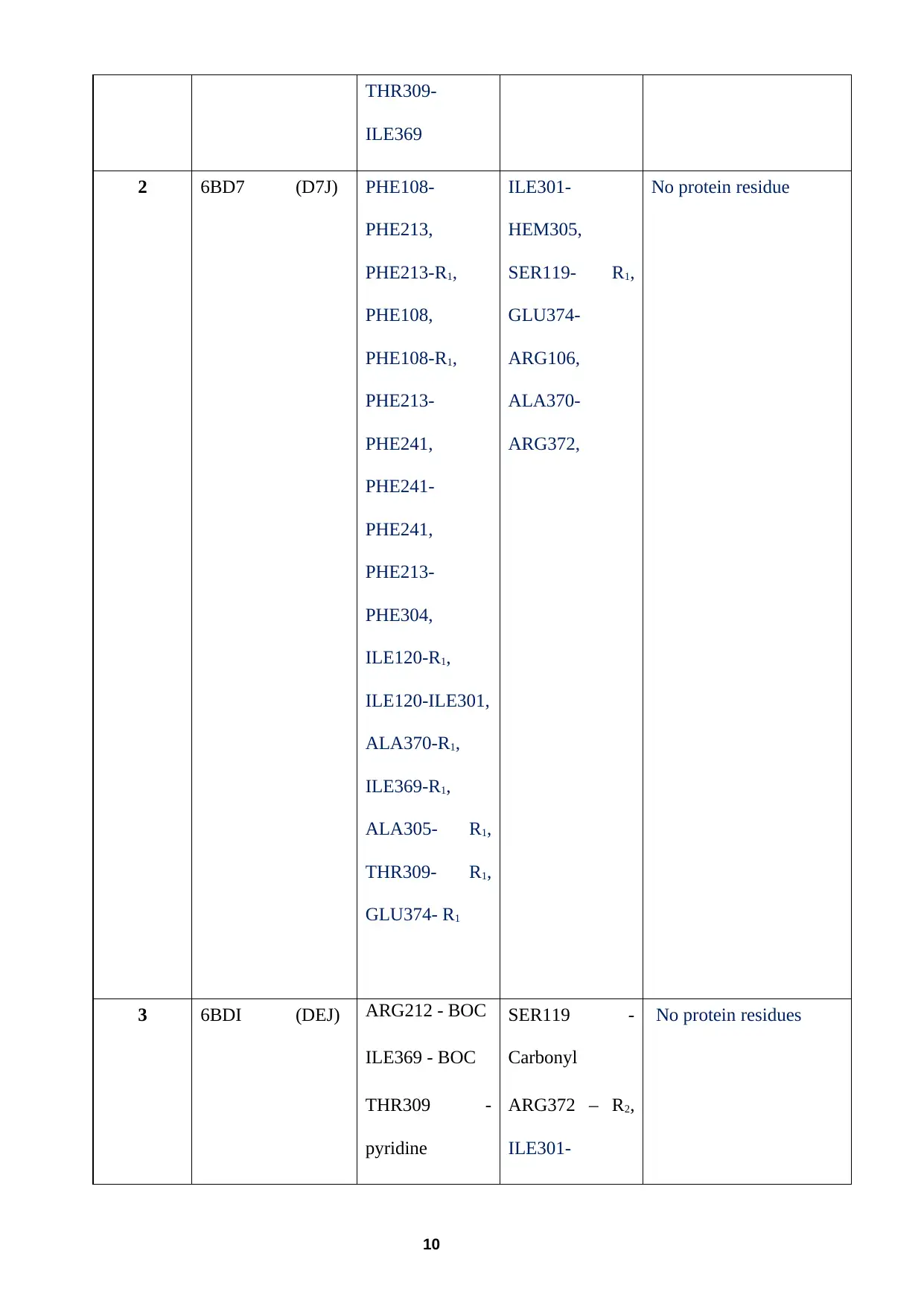
THR309-
ILE369
2 6BD7 (D7J) PHE108-
PHE213,
PHE213-R1,
PHE108,
PHE108-R1,
PHE213-
PHE241,
PHE241-
PHE241,
PHE213-
PHE304,
ILE120-R1,
ILE120-ILE301,
ALA370-R1,
ILE369-R1,
ALA305- R1,
THR309- R1,
GLU374- R1
ILE301-
HEM305,
SER119- R1,
GLU374-
ARG106,
ALA370-
ARG372,
No protein residue
3 6BDI (DEJ) ARG212 - BOC
ILE369 - BOC
THR309 -
pyridine
SER119 -
Carbonyl
ARG372 – R2,
ILE301-
No protein residues
10
ILE369
2 6BD7 (D7J) PHE108-
PHE213,
PHE213-R1,
PHE108,
PHE108-R1,
PHE213-
PHE241,
PHE241-
PHE241,
PHE213-
PHE304,
ILE120-R1,
ILE120-ILE301,
ALA370-R1,
ILE369-R1,
ALA305- R1,
THR309- R1,
GLU374- R1
ILE301-
HEM305,
SER119- R1,
GLU374-
ARG106,
ALA370-
ARG372,
No protein residue
3 6BDI (DEJ) ARG212 - BOC
ILE369 - BOC
THR309 -
pyridine
SER119 -
Carbonyl
ARG372 – R2,
ILE301-
No protein residues
10
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

ALA305 -
pyridine
and
PHE304 – R1
PHE215 – R1
or
PHE108 – R1
ILE120 – R1
ALA305,
ALA305-
THR309,
Instructions
Access the Protein Data Bank on the RCSB website (www.rcsb.org) and use the
search box to locate the entries for the crystal structure of human cytochrome
CYP3A4 bound to these compounds: 6BCZ (compound 1), 6BD7 (compound 2) and
6BDI (compound 3). Complete the entries in Table 2 for compounds 2.
You will need to use the new Ligand Viewer tool to explore each entry.
In the main Structure Summary page scroll down to the Small Molecules box and
select ‘Ligand Interaction’ for the appropriate entry in the Ligands table (see ligand
ID in Table 2 -above). Make sure that you have selected the ligand rather than the
haem group (HEM). This view shows the inhibitor molecule and the residues that
make up the overall binding pocket for the drug molecule.
Using this tool identify:
• the protein residues on the enzyme that are in hydrophobic contact with the
inhibitor molecule
11
pyridine
and
PHE304 – R1
PHE215 – R1
or
PHE108 – R1
ILE120 – R1
ALA305,
ALA305-
THR309,
Instructions
Access the Protein Data Bank on the RCSB website (www.rcsb.org) and use the
search box to locate the entries for the crystal structure of human cytochrome
CYP3A4 bound to these compounds: 6BCZ (compound 1), 6BD7 (compound 2) and
6BDI (compound 3). Complete the entries in Table 2 for compounds 2.
You will need to use the new Ligand Viewer tool to explore each entry.
In the main Structure Summary page scroll down to the Small Molecules box and
select ‘Ligand Interaction’ for the appropriate entry in the Ligands table (see ligand
ID in Table 2 -above). Make sure that you have selected the ligand rather than the
haem group (HEM). This view shows the inhibitor molecule and the residues that
make up the overall binding pocket for the drug molecule.
Using this tool identify:
• the protein residues on the enzyme that are in hydrophobic contact with the
inhibitor molecule
11

• any protein residues involved in π-interactions with the inhibitor which can be
either π−π stacking (marked in green) or π−cation (marked in orange).
• any protein residues involved in H−bonding with the inhibitor.
Enter the codes into Table 2.
(6 marks)
12
either π−π stacking (marked in green) or π−cation (marked in orange).
• any protein residues involved in H−bonding with the inhibitor.
Enter the codes into Table 2.
(6 marks)
12
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 15
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.
