Deep Learning Neural Networks for Brain Tumor Diagnosis: A Review
VerifiedAdded on 2024/07/01
|41
|7683
|343
Literature Review
AI Summary
This literature review explores the application of deep learning neural networks for medical image segmentation in brain tumor diagnosis. It examines several research papers focusing on techniques like integrating Conditional Random Fields (CRFs) with Fully Convolutional Neural Networks (FCNNs) to address challenges in MRI-based tumor segmentation. The review also covers 3D supervoxel-based learning methods for accurate tumor identification and automatic hierarchical tumor segmentation using Extremely Randomized Trees (ERT) to reduce manual segmentation efforts. The studies aim to improve the accuracy and efficiency of brain tumor diagnosis by leveraging advanced image processing and machine learning techniques, ultimately contributing to better treatment planning and patient outcomes. This document is contributed by a student and available on Desklib, a platform that offers study tools and solved assignments for students.
Literature Review (Secondary Research) Template
Student Name & CSU ID
Project Topic Title Deep learning neural networks for medical image segmentation of brain tumor diagnosis
Version 1.0 _ Week 1 (5 Journal Papers from CSU Library)
1
Reference in APA format that will be in
'Reference List'
Zhao, X., Wu, Y., Song, G., Li, Z., Zhang, Y., & Fan, Y. (2018). A deep learning model integrating FCNNs and
CRFs for brain tumor segmentation. Medical image analysis, Vol. 43, pp. 98-111.
Citation that will be in the content (Zhao, et. al., 2018)
URL of the Reference Level of Journal (Q1, Q2, …Qn) Keywords in this Reference
https://ac-els-cdn-
com.ezproxy.csu.edu.au/
S136184151730141X/1-s2.0-
S136184151730141X-main.pdf?
_tid=2da61e1d-7961-4740-9761-
0599e1e83c8b&acdnat=1532757075_b987
58be0e1f74b440498df61796621d
Q1 Conditional random fields, Brain tumour segmentation, Deep
learning, Fully convolutional neural networks
The Name of the Current Solution
(Technique/ Method/ Scheme/
Algorithm/ Model/ Tool/ Framework/ ...
etc )
The Goal (Objective) of this Solution &
What is the Problem that need to be solved
What are the components of it?
Technique/Algorithm name: Integrating
Conditional Random Fields (CRFs) and
Fully Conventional Neural Networks
(FCNNs)
Problem: The efforts have been done for
developing automatic or semi-automatic brain
tumour segmentation techniques and the
preferred method is MRI. However, there are
few challenges of using MRI’s such as gliomas
Voting based fusion method
Image patches
Image Slices
Recurrent Neural Networks (RNNs)
TC1 and TC2 scans
1
Student Name & CSU ID
Project Topic Title Deep learning neural networks for medical image segmentation of brain tumor diagnosis
Version 1.0 _ Week 1 (5 Journal Papers from CSU Library)
1
Reference in APA format that will be in
'Reference List'
Zhao, X., Wu, Y., Song, G., Li, Z., Zhang, Y., & Fan, Y. (2018). A deep learning model integrating FCNNs and
CRFs for brain tumor segmentation. Medical image analysis, Vol. 43, pp. 98-111.
Citation that will be in the content (Zhao, et. al., 2018)
URL of the Reference Level of Journal (Q1, Q2, …Qn) Keywords in this Reference
https://ac-els-cdn-
com.ezproxy.csu.edu.au/
S136184151730141X/1-s2.0-
S136184151730141X-main.pdf?
_tid=2da61e1d-7961-4740-9761-
0599e1e83c8b&acdnat=1532757075_b987
58be0e1f74b440498df61796621d
Q1 Conditional random fields, Brain tumour segmentation, Deep
learning, Fully convolutional neural networks
The Name of the Current Solution
(Technique/ Method/ Scheme/
Algorithm/ Model/ Tool/ Framework/ ...
etc )
The Goal (Objective) of this Solution &
What is the Problem that need to be solved
What are the components of it?
Technique/Algorithm name: Integrating
Conditional Random Fields (CRFs) and
Fully Conventional Neural Networks
(FCNNs)
Problem: The efforts have been done for
developing automatic or semi-automatic brain
tumour segmentation techniques and the
preferred method is MRI. However, there are
few challenges of using MRI’s such as gliomas
Voting based fusion method
Image patches
Image Slices
Recurrent Neural Networks (RNNs)
TC1 and TC2 scans
1
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
Tools: 2D image patches, Image slices,
voting based fusion method, BRATS
Applied Area: Brain tumour diagnosis
treatment and planning
may appear similarly to gliosis and stroke in
MRI
Goal: The aim is to develop a model based on
deep learning based segmentation model by
using 2D image patches.
The Process (Mechanism) of this Work; The process steps of the Technique/system
Process Steps Advantage (Purpose of this step) Disadvantage (Limitation/Challenge)
1 Imaging Data:
The first step is to obtain the imaging data from
BRATS 2015, BRATS 2016
It helps to collect basic information about the
MRI results that scans diagnosed patients with
gliomas
Only small volume of data is collected for
analysis
2 Brain tumour segmentation using FCNNs:
In this, the extracted image patches are
categorised by trained CNNs.
It reduces the issue of sample imbalance at the
testing stage
The relationship among the patches can be
lost while using patch-based segmentation
technique
3 Pre-processing of imaging data:
Using robust intensity normalisation technique
in order to compare the MRI scans and using
N41TK for correcting MRI data
By correcting the bias in the MRI data,
accurate results are obtained.
The results may not be accurate if the data is
incorrectly modified.
4 Comparing the results:
The results obtained are compared with the MRI
data for checking the accuracy
It helps to ascertain the viability of using the
proposed method
NA
Validation Criteria (Measurement Criteria)
2
voting based fusion method, BRATS
Applied Area: Brain tumour diagnosis
treatment and planning
may appear similarly to gliosis and stroke in
MRI
Goal: The aim is to develop a model based on
deep learning based segmentation model by
using 2D image patches.
The Process (Mechanism) of this Work; The process steps of the Technique/system
Process Steps Advantage (Purpose of this step) Disadvantage (Limitation/Challenge)
1 Imaging Data:
The first step is to obtain the imaging data from
BRATS 2015, BRATS 2016
It helps to collect basic information about the
MRI results that scans diagnosed patients with
gliomas
Only small volume of data is collected for
analysis
2 Brain tumour segmentation using FCNNs:
In this, the extracted image patches are
categorised by trained CNNs.
It reduces the issue of sample imbalance at the
testing stage
The relationship among the patches can be
lost while using patch-based segmentation
technique
3 Pre-processing of imaging data:
Using robust intensity normalisation technique
in order to compare the MRI scans and using
N41TK for correcting MRI data
By correcting the bias in the MRI data,
accurate results are obtained.
The results may not be accurate if the data is
incorrectly modified.
4 Comparing the results:
The results obtained are compared with the MRI
data for checking the accuracy
It helps to ascertain the viability of using the
proposed method
NA
Validation Criteria (Measurement Criteria)
2
Dependent Variable Independent Variable
Brain tumour segmentation Voxel’s label
Calculation of standard deviation Collected MRI scans
Input and Output Critical Thinking: Feature of this work, and
Why (Justify)
Critical Thinking: Limitations of the research
current solution, and Why (Justify)
Input (Data) Output (View)
Three segmentation
models are trained by
using 2D image
patches and
segmenting the brain
tumours using voting
based fusion method.
The results indicate
that the proposed
methods eliminate the
bias of MRI scan and
increase the efficiency
of the diagnosis.
The proposed method of training FCNNs and CRFs
helps to differentiate the gliosis and gliomas which
helps to provide appropriate diagnosis for cancer
patients.
The major drawback of applying this technique is
that the segmentation performance of the trained
network gets affected because the numbers of
pixels varies according to their class in the image
slices.
(Describe the research/current solution) Evaluation Criteria How this research/current solution is valuable
for your project
The current solution that is training FCNNs and
CRFs by using 2D image patches and segmenting
the images obtained from MRI scans.
The accuracy of the results obtained can be
evaluated from examining the scans which provides
appropriate analysis of the status of brain tumour.
There is a growing trend for using advanced
techniques for providing accurate diagnosis for
cancer. This research helps to understand the
techniques that can be used for developing future
technology in medical care.
Diagram/Flowchart
3
Brain tumour segmentation Voxel’s label
Calculation of standard deviation Collected MRI scans
Input and Output Critical Thinking: Feature of this work, and
Why (Justify)
Critical Thinking: Limitations of the research
current solution, and Why (Justify)
Input (Data) Output (View)
Three segmentation
models are trained by
using 2D image
patches and
segmenting the brain
tumours using voting
based fusion method.
The results indicate
that the proposed
methods eliminate the
bias of MRI scan and
increase the efficiency
of the diagnosis.
The proposed method of training FCNNs and CRFs
helps to differentiate the gliosis and gliomas which
helps to provide appropriate diagnosis for cancer
patients.
The major drawback of applying this technique is
that the segmentation performance of the trained
network gets affected because the numbers of
pixels varies according to their class in the image
slices.
(Describe the research/current solution) Evaluation Criteria How this research/current solution is valuable
for your project
The current solution that is training FCNNs and
CRFs by using 2D image patches and segmenting
the images obtained from MRI scans.
The accuracy of the results obtained can be
evaluated from examining the scans which provides
appropriate analysis of the status of brain tumour.
There is a growing trend for using advanced
techniques for providing accurate diagnosis for
cancer. This research helps to understand the
techniques that can be used for developing future
technology in medical care.
Diagram/Flowchart
3
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
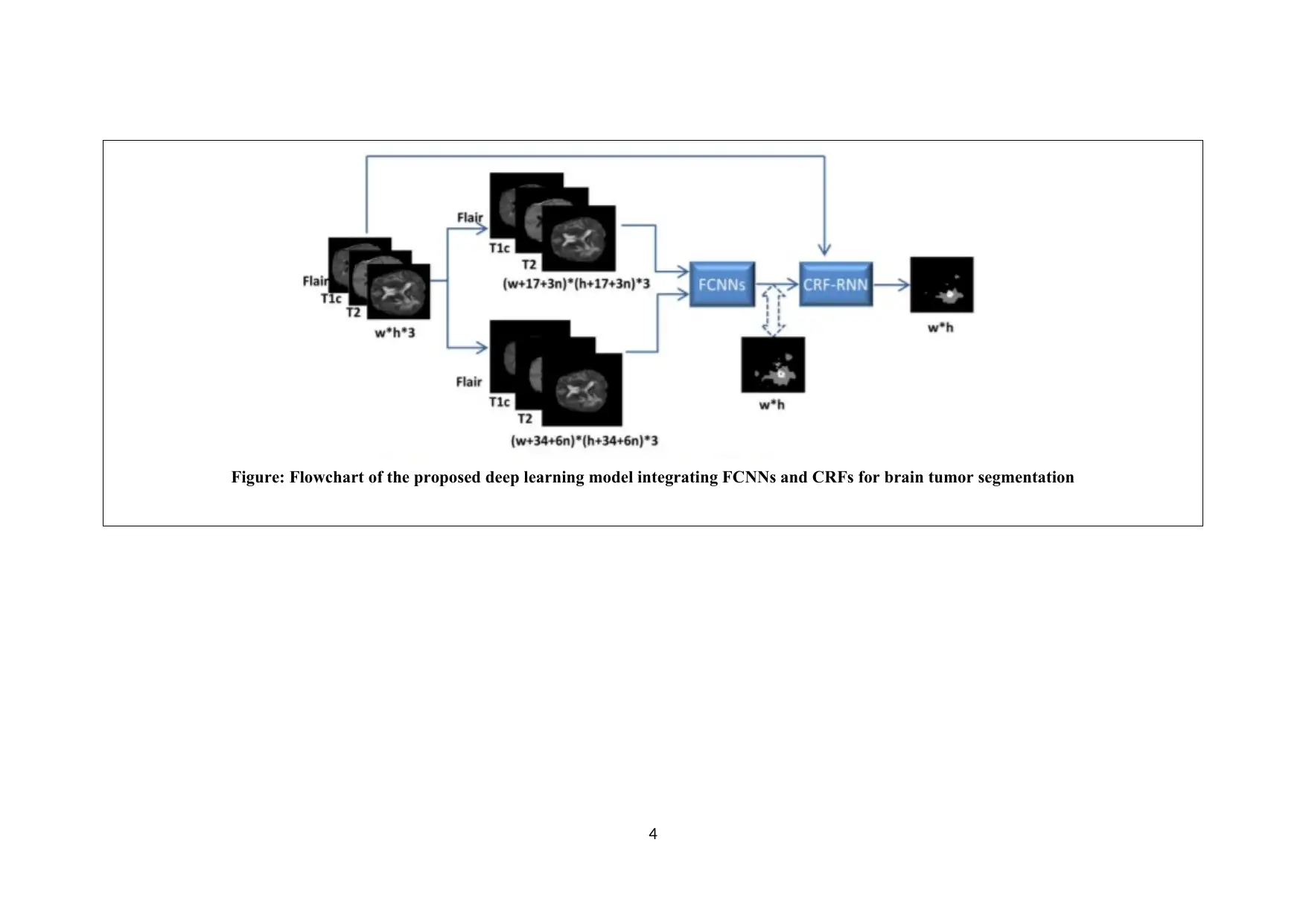
Figure: Flowchart of the proposed deep learning model integrating FCNNs and CRFs for brain tumor segmentation
4
4
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
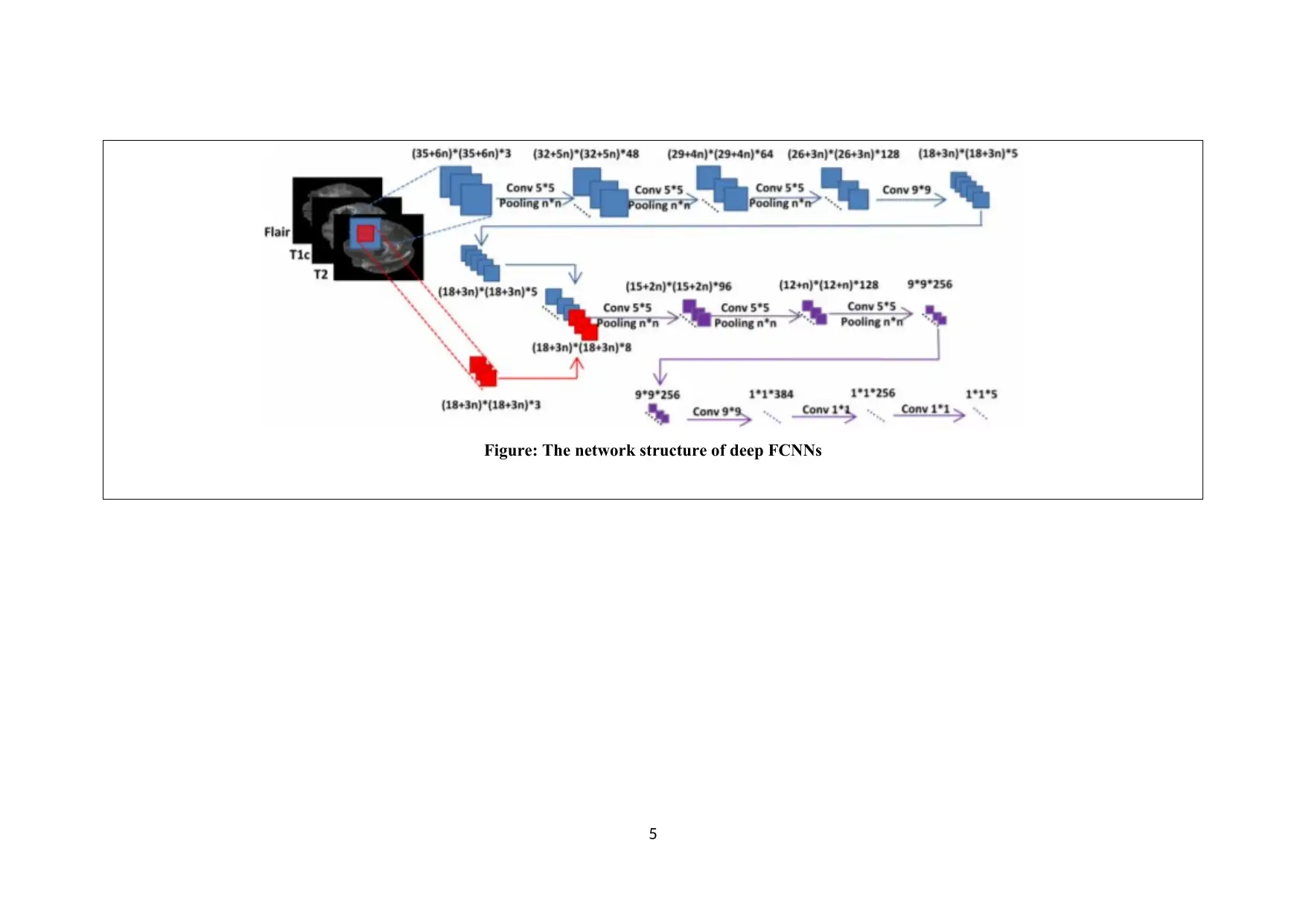
Figure: The network structure of deep FCNNs
5
5
2
Reference in APA format that will be in
'Reference List'
Soltaninejad, M., Yang, G., Lambrou, T., Allinson, N., Jones, T. L., Barrick, T. R., Howe, F.A., & Ye, X.
(2018). Supervised learning based multimodal MRI brain tumour segmentation using texture features from
supervoxels. Computer methods and programs in biomedicine, Vol. 157, pp. 69-84.
Citation that will be in the content (Soltaninejad, et. al., 2018)
URL of the Reference Level of Journal (Q1, Q2, …Qn) Keywords in this Reference
https://ac-els-cdn-
com.ezproxy.csu.edu.au/
S016926071731355X/1-s2.0-
S016926071731355X-main.pdf?
_tid=4a4dd16b-bfb6-41e4-b262-
b20025595ecb&acdnat=1532759931_766b
835216f08196959681dfbbd7ba82
Q1 Diffusion tensor imaging, Supervoxel, Brain tumour
segmentation, Multimodal MRI
The Name of the Current Solution
(Technique/ Method/ Scheme/
Algorithm/ Model/ Tool/ Framework/ ...
etc )
The Goal (Objective) of this Solution &
What is the Problem that need to be solved
What are the components of it?
Technique/Algorithm name: 3D
supervoxel based learning method
Tools: Generating supervoxels, Gabor
Filters, random forests (RF) classifier
Applied Area: Brain tumour segmentation
Problem: Because of numerous types of
tumours types, is becomes difficult for
segmenting the brain tumour in MRI.
Goal: The aim is to develop a 3D supervoxel
based learning method in order to accurately
identify the type of tumour after conducting
MRI scans.
Multimodal MRI images
Isotropic component
Anisotropic component
Diffusion Tensor Imaging (DTI)
Balanced Error Rate
6
Reference in APA format that will be in
'Reference List'
Soltaninejad, M., Yang, G., Lambrou, T., Allinson, N., Jones, T. L., Barrick, T. R., Howe, F.A., & Ye, X.
(2018). Supervised learning based multimodal MRI brain tumour segmentation using texture features from
supervoxels. Computer methods and programs in biomedicine, Vol. 157, pp. 69-84.
Citation that will be in the content (Soltaninejad, et. al., 2018)
URL of the Reference Level of Journal (Q1, Q2, …Qn) Keywords in this Reference
https://ac-els-cdn-
com.ezproxy.csu.edu.au/
S016926071731355X/1-s2.0-
S016926071731355X-main.pdf?
_tid=4a4dd16b-bfb6-41e4-b262-
b20025595ecb&acdnat=1532759931_766b
835216f08196959681dfbbd7ba82
Q1 Diffusion tensor imaging, Supervoxel, Brain tumour
segmentation, Multimodal MRI
The Name of the Current Solution
(Technique/ Method/ Scheme/
Algorithm/ Model/ Tool/ Framework/ ...
etc )
The Goal (Objective) of this Solution &
What is the Problem that need to be solved
What are the components of it?
Technique/Algorithm name: 3D
supervoxel based learning method
Tools: Generating supervoxels, Gabor
Filters, random forests (RF) classifier
Applied Area: Brain tumour segmentation
Problem: Because of numerous types of
tumours types, is becomes difficult for
segmenting the brain tumour in MRI.
Goal: The aim is to develop a 3D supervoxel
based learning method in order to accurately
identify the type of tumour after conducting
MRI scans.
Multimodal MRI images
Isotropic component
Anisotropic component
Diffusion Tensor Imaging (DTI)
Balanced Error Rate
6
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
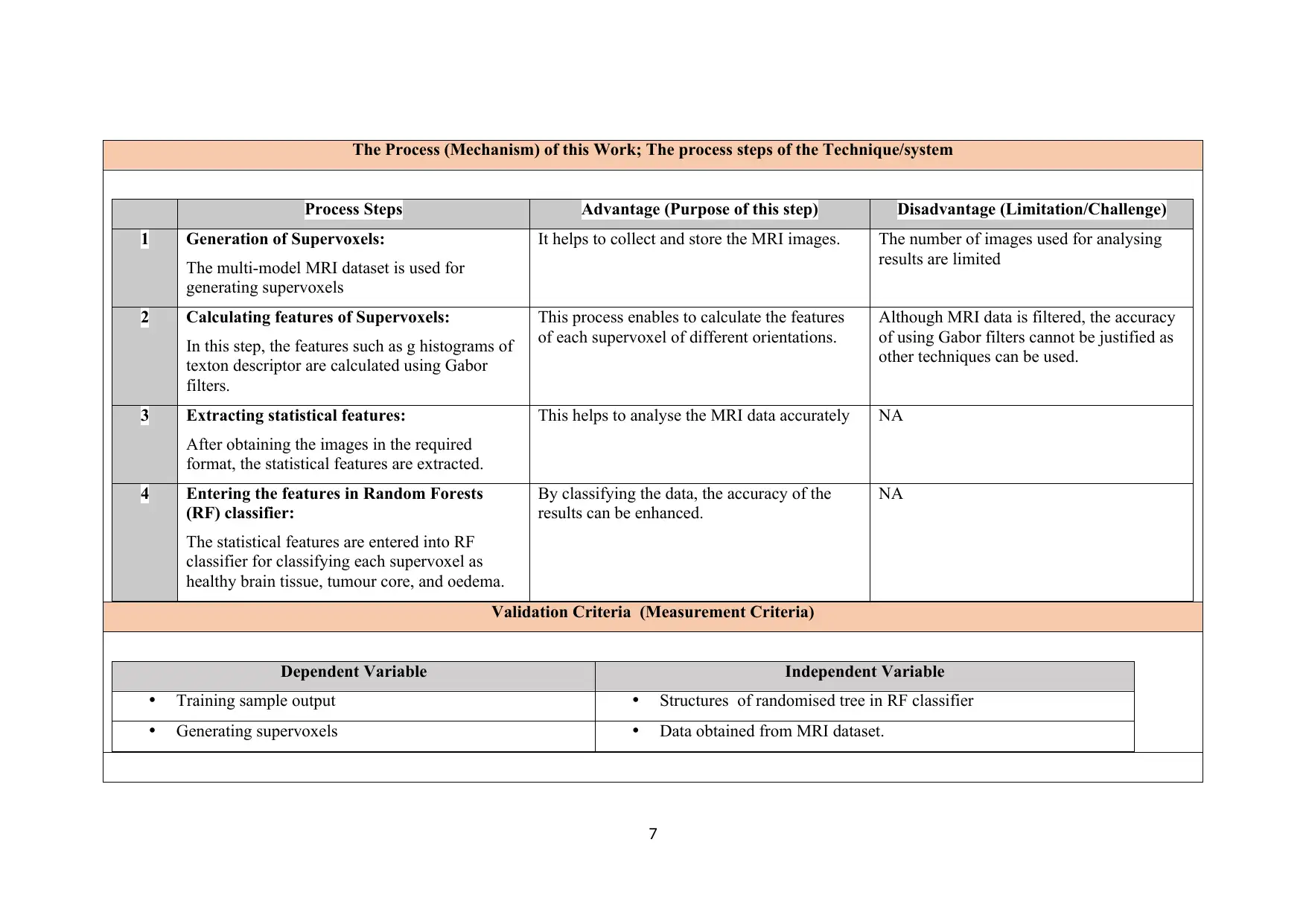
The Process (Mechanism) of this Work; The process steps of the Technique/system
Process Steps Advantage (Purpose of this step) Disadvantage (Limitation/Challenge)
1 Generation of Supervoxels:
The multi-model MRI dataset is used for
generating supervoxels
It helps to collect and store the MRI images. The number of images used for analysing
results are limited
2 Calculating features of Supervoxels:
In this step, the features such as g histograms of
texton descriptor are calculated using Gabor
filters.
This process enables to calculate the features
of each supervoxel of different orientations.
Although MRI data is filtered, the accuracy
of using Gabor filters cannot be justified as
other techniques can be used.
3 Extracting statistical features:
After obtaining the images in the required
format, the statistical features are extracted.
This helps to analyse the MRI data accurately NA
4 Entering the features in Random Forests
(RF) classifier:
The statistical features are entered into RF
classifier for classifying each supervoxel as
healthy brain tissue, tumour core, and oedema.
By classifying the data, the accuracy of the
results can be enhanced.
NA
Validation Criteria (Measurement Criteria)
Dependent Variable Independent Variable
Training sample output Structures of randomised tree in RF classifier
Generating supervoxels Data obtained from MRI dataset.
7
Process Steps Advantage (Purpose of this step) Disadvantage (Limitation/Challenge)
1 Generation of Supervoxels:
The multi-model MRI dataset is used for
generating supervoxels
It helps to collect and store the MRI images. The number of images used for analysing
results are limited
2 Calculating features of Supervoxels:
In this step, the features such as g histograms of
texton descriptor are calculated using Gabor
filters.
This process enables to calculate the features
of each supervoxel of different orientations.
Although MRI data is filtered, the accuracy
of using Gabor filters cannot be justified as
other techniques can be used.
3 Extracting statistical features:
After obtaining the images in the required
format, the statistical features are extracted.
This helps to analyse the MRI data accurately NA
4 Entering the features in Random Forests
(RF) classifier:
The statistical features are entered into RF
classifier for classifying each supervoxel as
healthy brain tissue, tumour core, and oedema.
By classifying the data, the accuracy of the
results can be enhanced.
NA
Validation Criteria (Measurement Criteria)
Dependent Variable Independent Variable
Training sample output Structures of randomised tree in RF classifier
Generating supervoxels Data obtained from MRI dataset.
7
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
Input and Output Critical Thinking: Feature of this work, and
Why (Justify)
Critical Thinking: Limitations of the research
current solution, and Why (Justify)
Input (Data) Output (View)
The MRI images are
collected for analysis
and supervoxels are
generated. Before
entering the data into
RF classifier, Gabor
Filters are used for
classifying the data.
The results obtained
by performing the
tests indicate that the
images provide
accurate results as to
the condition of the
tumour.
With the increasing number of cases related to brain
tumour, it is essential for developing a technology
which helps to accurately identify the tumours for
providing appropriate diagnosis.
The major drawback of using supervoxel is that
there is a minimum criterion that has to be fulfilled
with regards to its parameters.
(Describe the research/current solution) Evaluation Criteria How this research/current solution is valuable
for your project
The proposed 3D supervoxel based learning
method helps to eliminate the issues of MRI
scans. This is essential for ascertaining the actual
nature of tumour.
The MRI images are accurately classified as healthy
brain tissue, tumour core, and oedema which is
helpful while designing the type of diagnosis that is
to be provided to the patients.
3D supervoxel based learning method is a solution
for the issues currently being faced related to MRI
scans. This helps to develop an effective neural
networks and image processing technique.
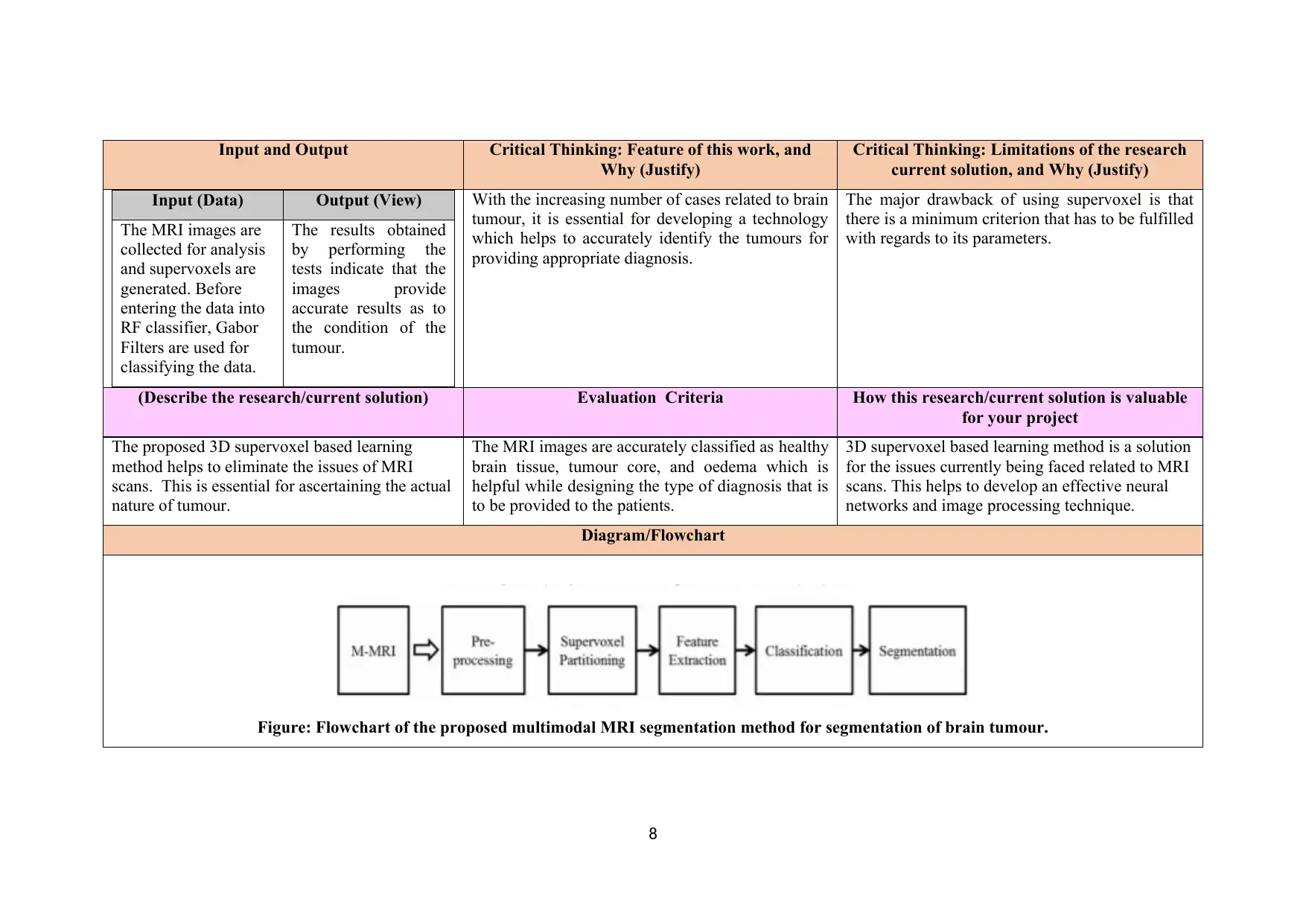
Diagram/Flowchart
Figure: Flowchart of the proposed multimodal MRI segmentation method for segmentation of brain tumour.
8
Why (Justify)
Critical Thinking: Limitations of the research
current solution, and Why (Justify)
Input (Data) Output (View)
The MRI images are
collected for analysis
and supervoxels are
generated. Before
entering the data into
RF classifier, Gabor
Filters are used for
classifying the data.
The results obtained
by performing the
tests indicate that the
images provide
accurate results as to
the condition of the
tumour.
With the increasing number of cases related to brain
tumour, it is essential for developing a technology
which helps to accurately identify the tumours for
providing appropriate diagnosis.
The major drawback of using supervoxel is that
there is a minimum criterion that has to be fulfilled
with regards to its parameters.
(Describe the research/current solution) Evaluation Criteria How this research/current solution is valuable
for your project
The proposed 3D supervoxel based learning
method helps to eliminate the issues of MRI
scans. This is essential for ascertaining the actual
nature of tumour.
The MRI images are accurately classified as healthy
brain tissue, tumour core, and oedema which is
helpful while designing the type of diagnosis that is
to be provided to the patients.
3D supervoxel based learning method is a solution
for the issues currently being faced related to MRI
scans. This helps to develop an effective neural
networks and image processing technique.
Diagram/Flowchart
Figure: Flowchart of the proposed multimodal MRI segmentation method for segmentation of brain tumour.
8
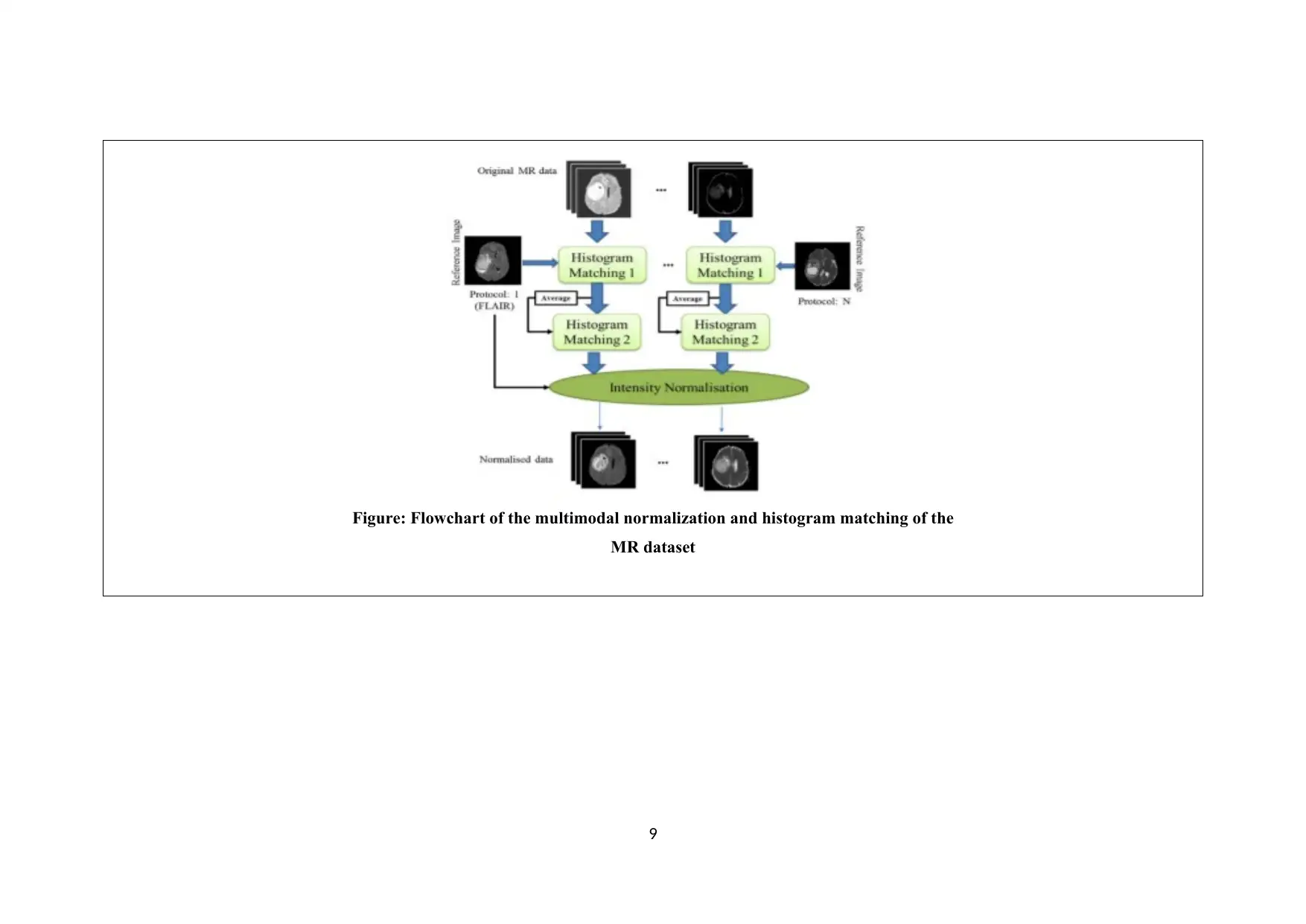
Figure: Flowchart of the multimodal normalization and histogram matching of the
MR dataset
9
MR dataset
9
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
3
Reference in APA format that will be in
'Reference List'
Pinto, A., Pereira, S., Rasteiro, D., & Silva, C. A. (2018). Hierarchical Brain Tumour Segmentation using
Extremely Randomized Trees. Pattern Recognition.
Citation that will be in the content (Pinto, et. al., 2018)
URL of the Reference Level of Journal (Q1, Q2, …Qn) Keywords in this Reference
https://ac-els-cdn-
com.ezproxy.csu.edu.au/
S0031320318301699/1-s2.0-
S0031320318301699-main.pdf?
_tid=6cec5e43-2f0c-46c9-941d-
276563fcb78d&acdnat=1532759868_53c8
b955cadbe0e1d6b5046a6a61f0c7
Q1 Magnetic resonance imaging, Extremely randomized trees,
Machine learning, Image segmentation
The Name of the Current Solution
(Technique/ Method/ Scheme/
Algorithm/ Model/ Tool/ Framework/ ...
etc )
The Goal (Objective) of this Solution &
What is the Problem that need to be solved
What are the components of it?
Technique/Algorithm name: Automatic
hierarchical brain tumour segmentation
using Extremely Randomized Trees (ERT)
Tools: Binary classification, morphological
refinement, N4ITK method, histogram
matching method
Applied Area: Brain Tumour
Segmentation
Problem: MRI is the commonly used method
for identifying gliomas. But this requires
manual segmentation which is difficult as well
as time-consuming task.
Goal: The aim is to develop an automatic
tumour segmentation technique using
Extremely Randomized Trees for accurate
analysis of the condition of tumour.
BRATS 2013
Bias field distortion
Intensity normalisation
Data augmentation
2-dimensional patches
Tumour detection
10
Reference in APA format that will be in
'Reference List'
Pinto, A., Pereira, S., Rasteiro, D., & Silva, C. A. (2018). Hierarchical Brain Tumour Segmentation using
Extremely Randomized Trees. Pattern Recognition.
Citation that will be in the content (Pinto, et. al., 2018)
URL of the Reference Level of Journal (Q1, Q2, …Qn) Keywords in this Reference
https://ac-els-cdn-
com.ezproxy.csu.edu.au/
S0031320318301699/1-s2.0-
S0031320318301699-main.pdf?
_tid=6cec5e43-2f0c-46c9-941d-
276563fcb78d&acdnat=1532759868_53c8
b955cadbe0e1d6b5046a6a61f0c7
Q1 Magnetic resonance imaging, Extremely randomized trees,
Machine learning, Image segmentation
The Name of the Current Solution
(Technique/ Method/ Scheme/
Algorithm/ Model/ Tool/ Framework/ ...
etc )
The Goal (Objective) of this Solution &
What is the Problem that need to be solved
What are the components of it?
Technique/Algorithm name: Automatic
hierarchical brain tumour segmentation
using Extremely Randomized Trees (ERT)
Tools: Binary classification, morphological
refinement, N4ITK method, histogram
matching method
Applied Area: Brain Tumour
Segmentation
Problem: MRI is the commonly used method
for identifying gliomas. But this requires
manual segmentation which is difficult as well
as time-consuming task.
Goal: The aim is to develop an automatic
tumour segmentation technique using
Extremely Randomized Trees for accurate
analysis of the condition of tumour.
BRATS 2013
Bias field distortion
Intensity normalisation
Data augmentation
2-dimensional patches
Tumour detection
10
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
The Process (Mechanism) of this Work; The process steps of the Technique/system
Process Steps Advantage (Purpose of this step) Disadvantage (Limitation/Challenge)
1 Collecting the data:
The MRI images are collected for analysing the
accuracy of using Extremely Randomized Trees.
It helps to ascertain the accuracy that can be
achieved by using the proposed technique.
A large sample has to be analysed for
accurate analysis of the data.
2 Correcting the image bias:
The imperfections in the MRI images are
corrected using N4ITK method
This minimises the distortions in the images
and enhances the accuracy of the results.
Although the imperfections are reduced, it
does not help to completely eliminate wrong
data.
3 Intensity Normalisation:
The histogram matching method is used during
this stage for ensuring identical intensity
distribution of the same sequence
This enables reducing the differences between
the images
NA
4 Analysis of the data:
The results are compared with existing
techniques used such as T1-weighted, and d
Fluid Attenuation Inversion Recovery
This helps to understand the viability of using
this technique.
NA
Validation Criteria (Measurement Criteria)
Dependent Variable Independent Variable
Tumour classifications Complexity of the tumour
Calculation of intensities intensity variability of the percentiles of the training images
Input and Output Critical Thinking: Feature of this work, and Critical Thinking: Limitations of the research
11
Process Steps Advantage (Purpose of this step) Disadvantage (Limitation/Challenge)
1 Collecting the data:
The MRI images are collected for analysing the
accuracy of using Extremely Randomized Trees.
It helps to ascertain the accuracy that can be
achieved by using the proposed technique.
A large sample has to be analysed for
accurate analysis of the data.
2 Correcting the image bias:
The imperfections in the MRI images are
corrected using N4ITK method
This minimises the distortions in the images
and enhances the accuracy of the results.
Although the imperfections are reduced, it
does not help to completely eliminate wrong
data.
3 Intensity Normalisation:
The histogram matching method is used during
this stage for ensuring identical intensity
distribution of the same sequence
This enables reducing the differences between
the images
NA
4 Analysis of the data:
The results are compared with existing
techniques used such as T1-weighted, and d
Fluid Attenuation Inversion Recovery
This helps to understand the viability of using
this technique.
NA
Validation Criteria (Measurement Criteria)
Dependent Variable Independent Variable
Tumour classifications Complexity of the tumour
Calculation of intensities intensity variability of the percentiles of the training images
Input and Output Critical Thinking: Feature of this work, and Critical Thinking: Limitations of the research
11
Why (Justify) current solution, and Why (Justify)
Input (Data) Output (View)
For arriving at the
desired results, the
MRI scan images are
collected. The bias in
the images are
normalised before
entering the same into
Random Forest (RF
classifier).
The output is verified
by considering the
metrics, Positive
Predictive Value,
Dice Similarity
Coefficient and
Sensitivity.
The proposed technique helps to analyse the types
of tumour by eliminating biases. Hence, accuracy of
the diagnosis is ensured.
The intensity range of the necrosis tissues would
have been shifted affecting the training of
classifier.
(Describe the research/current solution) Evaluation Criteria How this research/current solution is valuable
for your project
The use of the proposed technique reduces the
manual segmentation of tumour tissues thereby
enhancing the accuracy of the results.
The images are clearer and the affected brain area is
highlighted along with corresponding tissues.
The proposed research will help to develop a
novel technique which can be used for developing
advanced methods for detecting brain tumours.
Diagram/Flowchart
12
Input (Data) Output (View)
For arriving at the
desired results, the
MRI scan images are
collected. The bias in
the images are
normalised before
entering the same into
Random Forest (RF
classifier).
The output is verified
by considering the
metrics, Positive
Predictive Value,
Dice Similarity
Coefficient and
Sensitivity.
The proposed technique helps to analyse the types
of tumour by eliminating biases. Hence, accuracy of
the diagnosis is ensured.
The intensity range of the necrosis tissues would
have been shifted affecting the training of
classifier.
(Describe the research/current solution) Evaluation Criteria How this research/current solution is valuable
for your project
The use of the proposed technique reduces the
manual segmentation of tumour tissues thereby
enhancing the accuracy of the results.
The images are clearer and the affected brain area is
highlighted along with corresponding tissues.
The proposed research will help to develop a
novel technique which can be used for developing
advanced methods for detecting brain tumours.
Diagram/Flowchart
12
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 41
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.