Comprehensive Report: Determination of Proteomics in Grape Skin
VerifiedAdded on 2020/04/07
|15
|3705
|161
Report
AI Summary
This report provides a comprehensive overview of the determination of proteomics in grape skin. It begins with an introduction to proteomics and a literature review on grape skin proteins, emphasizing their role in the ripening and development of grapes. The report details various sample preparation methods, including disruption techniques using chaotropes and surfactants, and methods for alkylation and reduction of cysteine. It then explores fractionation methods to remove contaminants and separate proteins, followed by a discussion of analytical methods such as comparative 2-DE, MS analysis, and SDS-PAGE. The report highlights the advantages of 1D PAGE for protein measurement and concludes with a bioinformatics method of analysis and a conclusion summarizing the findings. The report also includes a flowchart of the proteome analysis strategy and a table illustrating the interactions between and within the proteins, method used during the process of disruption, and the energy needed during the disruption of the interaction of the proteins in the grape skin.

Report on the determination of proteomics on rape skin 1
REPORT ON THE DETERMINATION OF PROTEOMICS OF GRAPE SKIN
A Report Paper on Proteins By
Student’s Name
Name of the Professor
Institutional Affiliation
City/State
Year/Month/Day
REPORT ON THE DETERMINATION OF PROTEOMICS OF GRAPE SKIN
A Report Paper on Proteins By
Student’s Name
Name of the Professor
Institutional Affiliation
City/State
Year/Month/Day
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Report on the determination of proteomics on rape skin 2
TABLE OF CONTENT
Item Page
INTRODUCTION……………………………………………………………….…….3
LITERATURE REVIEW……………………………………………………………..3
Proteins……………….………………………………………………………4
Chaotropes………………………………………………………………….....5
SAMPLE PREPARATION METHODS…………………………………………….5
Surfactants…………….……………………………………………………...6
Alkylation and reduction of cysteine…….…………………………………...6
FRACTIONATION METHODS .....................................................................….......7
METHODS OF ANALYSIS……………………………….……….………………..8
1. Comparative 2-DE…………………………………..……….……………….8
2. MS Analysis………………………………………..….……………………..8
3. SDS-PAGE……………………………………………………………………9
SDS PAGE……………….………………………………………….………………10
Mass Spectrometry Method………………………………………………….11
BIOINFORMATICS METHOD OF ANALYSIS………………………………….12
CONCLUSION………………………………………………………………………13
BIBLIOGRAPHY……………………………………………………………………14
TABLE OF CONTENT
Item Page
INTRODUCTION……………………………………………………………….…….3
LITERATURE REVIEW……………………………………………………………..3
Proteins……………….………………………………………………………4
Chaotropes………………………………………………………………….....5
SAMPLE PREPARATION METHODS…………………………………………….5
Surfactants…………….……………………………………………………...6
Alkylation and reduction of cysteine…….…………………………………...6
FRACTIONATION METHODS .....................................................................….......7
METHODS OF ANALYSIS……………………………….……….………………..8
1. Comparative 2-DE…………………………………..……….……………….8
2. MS Analysis………………………………………..….……………………..8
3. SDS-PAGE……………………………………………………………………9
SDS PAGE……………….………………………………………….………………10
Mass Spectrometry Method………………………………………………….11
BIOINFORMATICS METHOD OF ANALYSIS………………………………….12
CONCLUSION………………………………………………………………………13
BIBLIOGRAPHY……………………………………………………………………14

Report on the determination of proteomics on rape skin 3
TASK 2
Title: Report on Proteomics
Introduction
The biological term proteomics is used in describing the study of a full set of
proteome or proteins which are expressed at a time provided in an organism, cell, organ or
tissue. The current proteomics includes the integration of an inclusive of tools of protein-
analytical information technologies and tools to reliably and quickly identify quantitative and
qualitative variations in proteins. This will include recognition of changed protein expression
related to the disease. This report also covers primarily for mass spectrometry, sample
separations, separations, and protein fractionations for protein characterization and
identification.
The structured program which is exploring facets of the data analysis and
experimental design have been undertaken and also the technologies of sample-preparation
which are practised exclusively for numerous types of samples such as mammalian fluids and
tissue, plants, and micro-organisms. There is need of independent research of literature that is
relevant to this topic. The practical section enables the understanding of the methodology and
logic underpinning preparation of sample, protein separation, mass spectrometry, and
fractionation of complex mixture.
Literature Review
In this report, the sample which was chosen was a grape skin which was analysed
through the determination of its proteome. The two stages of ripening of the grape skin were
analysed after a duration of two weeks apart. The samples were stored out of the refrigerator
to enable a process that is natural to take place. The skin of the grapes were chosen since the
tissues of the skin are present in the whole of the entire ripening and development of the
TASK 2
Title: Report on Proteomics
Introduction
The biological term proteomics is used in describing the study of a full set of
proteome or proteins which are expressed at a time provided in an organism, cell, organ or
tissue. The current proteomics includes the integration of an inclusive of tools of protein-
analytical information technologies and tools to reliably and quickly identify quantitative and
qualitative variations in proteins. This will include recognition of changed protein expression
related to the disease. This report also covers primarily for mass spectrometry, sample
separations, separations, and protein fractionations for protein characterization and
identification.
The structured program which is exploring facets of the data analysis and
experimental design have been undertaken and also the technologies of sample-preparation
which are practised exclusively for numerous types of samples such as mammalian fluids and
tissue, plants, and micro-organisms. There is need of independent research of literature that is
relevant to this topic. The practical section enables the understanding of the methodology and
logic underpinning preparation of sample, protein separation, mass spectrometry, and
fractionation of complex mixture.
Literature Review
In this report, the sample which was chosen was a grape skin which was analysed
through the determination of its proteome. The two stages of ripening of the grape skin were
analysed after a duration of two weeks apart. The samples were stored out of the refrigerator
to enable a process that is natural to take place. The skin of the grapes were chosen since the
tissues of the skin are present in the whole of the entire ripening and development of the
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Report on the determination of proteomics on rape skin 4
grape and also the skin acts as a critical physical barrier for the protection of the grapes from
threats which are external like external damages and pathogens (Akay, 2011, p. 156).
Proteins in Grape Skin
A protein is a chain of amino acids which are joined together by a band known as
peptide among every amino acid. There are twenty different types of amino acids which form
the major components of numerous proteins. Diverse amino acids have diverse functional
groups on the chains located on their sides. The functional groups may be different
chemically and co-operate with the functional groups on other different amino acids, such as
in the similar molecule of proteins and other diverse molecules of proteins. The resultant
compound normally determines the structure and shape of the protein molecules making up
the skin of grapes which are used in the report (Daoud, 2011, p. 268).
Sample Preparation Methods
Grape skin disruption and initial solubility during the analysis of its proteome
The disruption of the of the skin tissues in the initial stage of proteomic fractionation
strategy. This is done by homogenization of the tissues in the skin of the grapes by the use of
cryogenic grinding or Doust homogeniser using the frozen grape skin in a pestle and motor.
This requires being done swiftly in the presence of suitable inhibitors of protease to reduce
degradation of photolytic of the grape skin. This alteration does not break the cells of the
grape skin making the interactions and complexes of proteins to be preserved (Ferenc Darvas,
2016, p. 189).
In order to break and open the cells of the grape cells, mechanical strategies may be
incorporated such as ultrasonic disruptions by the use of pressure devices or probe sonicators,
cryogenic grinding by the use of bead mill, and thawing/freeze. The buffer selection utilized
at this stage to solubilise the proteins is critical since if the protein cannot be solubilised, then
grape and also the skin acts as a critical physical barrier for the protection of the grapes from
threats which are external like external damages and pathogens (Akay, 2011, p. 156).
Proteins in Grape Skin
A protein is a chain of amino acids which are joined together by a band known as
peptide among every amino acid. There are twenty different types of amino acids which form
the major components of numerous proteins. Diverse amino acids have diverse functional
groups on the chains located on their sides. The functional groups may be different
chemically and co-operate with the functional groups on other different amino acids, such as
in the similar molecule of proteins and other diverse molecules of proteins. The resultant
compound normally determines the structure and shape of the protein molecules making up
the skin of grapes which are used in the report (Daoud, 2011, p. 268).
Sample Preparation Methods
Grape skin disruption and initial solubility during the analysis of its proteome
The disruption of the of the skin tissues in the initial stage of proteomic fractionation
strategy. This is done by homogenization of the tissues in the skin of the grapes by the use of
cryogenic grinding or Doust homogeniser using the frozen grape skin in a pestle and motor.
This requires being done swiftly in the presence of suitable inhibitors of protease to reduce
degradation of photolytic of the grape skin. This alteration does not break the cells of the
grape skin making the interactions and complexes of proteins to be preserved (Ferenc Darvas,
2016, p. 189).
In order to break and open the cells of the grape cells, mechanical strategies may be
incorporated such as ultrasonic disruptions by the use of pressure devices or probe sonicators,
cryogenic grinding by the use of bead mill, and thawing/freeze. The buffer selection utilized
at this stage to solubilise the proteins is critical since if the protein cannot be solubilised, then
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
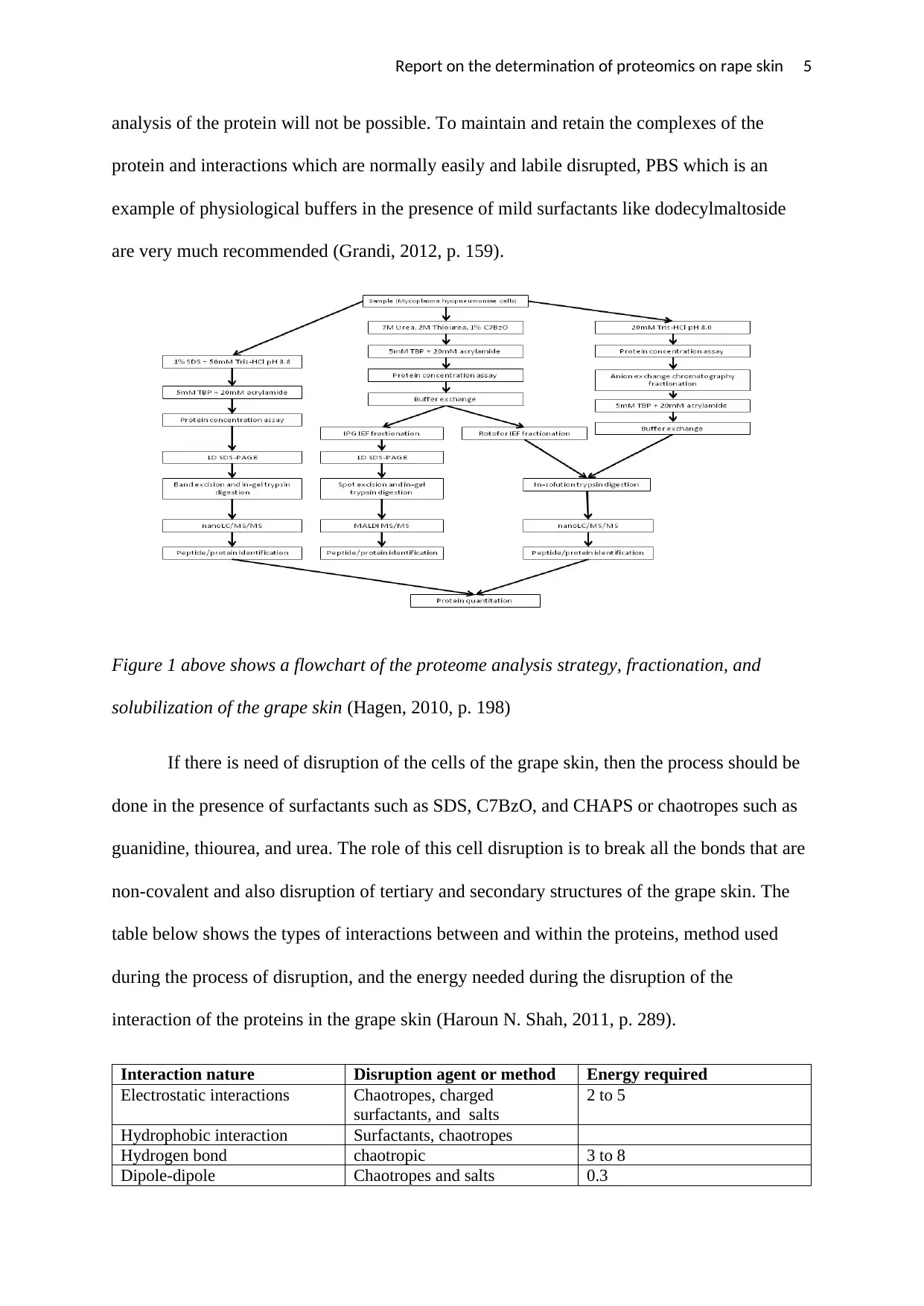
Report on the determination of proteomics on rape skin 5
analysis of the protein will not be possible. To maintain and retain the complexes of the
protein and interactions which are normally easily and labile disrupted, PBS which is an
example of physiological buffers in the presence of mild surfactants like dodecylmaltoside
are very much recommended (Grandi, 2012, p. 159).
Figure 1 above shows a flowchart of the proteome analysis strategy, fractionation, and
solubilization of the grape skin (Hagen, 2010, p. 198)
If there is need of disruption of the cells of the grape skin, then the process should be
done in the presence of surfactants such as SDS, C7BzO, and CHAPS or chaotropes such as
guanidine, thiourea, and urea. The role of this cell disruption is to break all the bonds that are
non-covalent and also disruption of tertiary and secondary structures of the grape skin. The
table below shows the types of interactions between and within the proteins, method used
during the process of disruption, and the energy needed during the disruption of the
interaction of the proteins in the grape skin (Haroun N. Shah, 2011, p. 289).
Interaction nature Disruption agent or method Energy required
Electrostatic interactions Chaotropes, charged
surfactants, and salts
2 to 5
Hydrophobic interaction Surfactants, chaotropes
Hydrogen bond chaotropic 3 to 8
Dipole-dipole Chaotropes and salts 0.3
analysis of the protein will not be possible. To maintain and retain the complexes of the
protein and interactions which are normally easily and labile disrupted, PBS which is an
example of physiological buffers in the presence of mild surfactants like dodecylmaltoside
are very much recommended (Grandi, 2012, p. 159).
Figure 1 above shows a flowchart of the proteome analysis strategy, fractionation, and
solubilization of the grape skin (Hagen, 2010, p. 198)
If there is need of disruption of the cells of the grape skin, then the process should be
done in the presence of surfactants such as SDS, C7BzO, and CHAPS or chaotropes such as
guanidine, thiourea, and urea. The role of this cell disruption is to break all the bonds that are
non-covalent and also disruption of tertiary and secondary structures of the grape skin. The
table below shows the types of interactions between and within the proteins, method used
during the process of disruption, and the energy needed during the disruption of the
interaction of the proteins in the grape skin (Haroun N. Shah, 2011, p. 289).
Interaction nature Disruption agent or method Energy required
Electrostatic interactions Chaotropes, charged
surfactants, and salts
2 to 5
Hydrophobic interaction Surfactants, chaotropes
Hydrogen bond chaotropic 3 to 8
Dipole-dipole Chaotropes and salts 0.3

Report on the determination of proteomics on rape skin 6
Van der Waals
Charge-dipole
Van der Waals
Chaotropes and salts 0.1
Disulphide bond Reduction 40
Chaotropes
The method of chaotropes function through disruption of the hydrogen bonds of the water at
the protein surface. When bonds between hydrogen and water join to the chaotropic instead
of the proteins, the proteins will unfold leading to the exposure of interior protein. The
chaotropic will then be removed from this solution since it is not needed through the use of
buffer exchange (Hondermarck, 2016, p. 189).
Surfactants
These are active surface agents which normally utilize compounds which are soluble in water
which minimizes the liquid's surface tension or minimize the tension of the interface between
a solid and a liquid or liquid and solid. The best solubilizing protein surfactant is the sodium
dodecyl sulphate and anionic surfactant (Ian Humphery-Smith, 2013, p. 178).
Alkylation and reduction of cysteine
The final step of full protein disruption in the grape skin is the reduction of bridges of
disulphide between alkylation and amino acids of the free thiol to inhibit the reformation of
disulphides. The DTT is supplemented and permitted to react for thirty minutes or more
followed by the addition of an agent of alkylation such as iodoacetamide (Joanna S. Albala,
2013, p. 248).
Fractionation Methods involved in Proteomic of grape skin
Removal of reagents and contaminants
Van der Waals
Charge-dipole
Van der Waals
Chaotropes and salts 0.1
Disulphide bond Reduction 40
Chaotropes
The method of chaotropes function through disruption of the hydrogen bonds of the water at
the protein surface. When bonds between hydrogen and water join to the chaotropic instead
of the proteins, the proteins will unfold leading to the exposure of interior protein. The
chaotropic will then be removed from this solution since it is not needed through the use of
buffer exchange (Hondermarck, 2016, p. 189).
Surfactants
These are active surface agents which normally utilize compounds which are soluble in water
which minimizes the liquid's surface tension or minimize the tension of the interface between
a solid and a liquid or liquid and solid. The best solubilizing protein surfactant is the sodium
dodecyl sulphate and anionic surfactant (Ian Humphery-Smith, 2013, p. 178).
Alkylation and reduction of cysteine
The final step of full protein disruption in the grape skin is the reduction of bridges of
disulphide between alkylation and amino acids of the free thiol to inhibit the reformation of
disulphides. The DTT is supplemented and permitted to react for thirty minutes or more
followed by the addition of an agent of alkylation such as iodoacetamide (Joanna S. Albala,
2013, p. 248).
Fractionation Methods involved in Proteomic of grape skin
Removal of reagents and contaminants
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Report on the determination of proteomics on rape skin 7
The techniques involved in a deduction of proteome complexity operate at their best
when there is specifically protein and chosen reagents existing. These reagents are important
in maintaining the solubility of the proteins during the technique of fractionation and precise
to the fractionation technique. Cells have a wide range of chemical compositions and not
only proteins alone and this will affect the fractionation process. The contaminants should be
eliminated or the technique of fractionation chosen initially which is not sensitive to the
presence of the contaminants (Joanna S. Albala, 2013, p. 358).
Buffers and Salts
High salt concentration or specific physiological buffers may affect in numerous ways
with chemical labelling, chromatography, and electrophoresis which involved SDC-PAGE
and isoelectric. The removal of buffer and salt can be done through the following meter of
conductivity: solid phase extraction, dialysis, precipitation, dilution, and buffer exchange by
the use of gel chromatography filtration (Joanna S. Albala, 2013, p. 187).
Removal of lipids: The surfactants which are present in the solution is enough to remove the
lipids which are soluble hence preventing interference. Lipids can also be removed through
precipitation or solvent extraction using acetone precipitation.
Removal of nucleic acids: These components can be removed through ultra-centrifugation
which huge nucleic acids may be are added to disrupt the interaction between nucleic acid
and protein, endonuclease where an enzyme is engineered to work in denaturing solution or
mechanical disruption where bead mills and ultrasonic probes are efficient at nucleic acids
that are shear (Jozef Samaj, 2013, p. 179).
Desalting of Grape skin
Desalting though the centrifugal process is a faster method of carrying out the gel-
filtration chromatography and also need a low-velocity centrifuge as compared to the system
The techniques involved in a deduction of proteome complexity operate at their best
when there is specifically protein and chosen reagents existing. These reagents are important
in maintaining the solubility of the proteins during the technique of fractionation and precise
to the fractionation technique. Cells have a wide range of chemical compositions and not
only proteins alone and this will affect the fractionation process. The contaminants should be
eliminated or the technique of fractionation chosen initially which is not sensitive to the
presence of the contaminants (Joanna S. Albala, 2013, p. 358).
Buffers and Salts
High salt concentration or specific physiological buffers may affect in numerous ways
with chemical labelling, chromatography, and electrophoresis which involved SDC-PAGE
and isoelectric. The removal of buffer and salt can be done through the following meter of
conductivity: solid phase extraction, dialysis, precipitation, dilution, and buffer exchange by
the use of gel chromatography filtration (Joanna S. Albala, 2013, p. 187).
Removal of lipids: The surfactants which are present in the solution is enough to remove the
lipids which are soluble hence preventing interference. Lipids can also be removed through
precipitation or solvent extraction using acetone precipitation.
Removal of nucleic acids: These components can be removed through ultra-centrifugation
which huge nucleic acids may be are added to disrupt the interaction between nucleic acid
and protein, endonuclease where an enzyme is engineered to work in denaturing solution or
mechanical disruption where bead mills and ultrasonic probes are efficient at nucleic acids
that are shear (Jozef Samaj, 2013, p. 179).
Desalting of Grape skin
Desalting though the centrifugal process is a faster method of carrying out the gel-
filtration chromatography and also need a low-velocity centrifuge as compared to the system
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
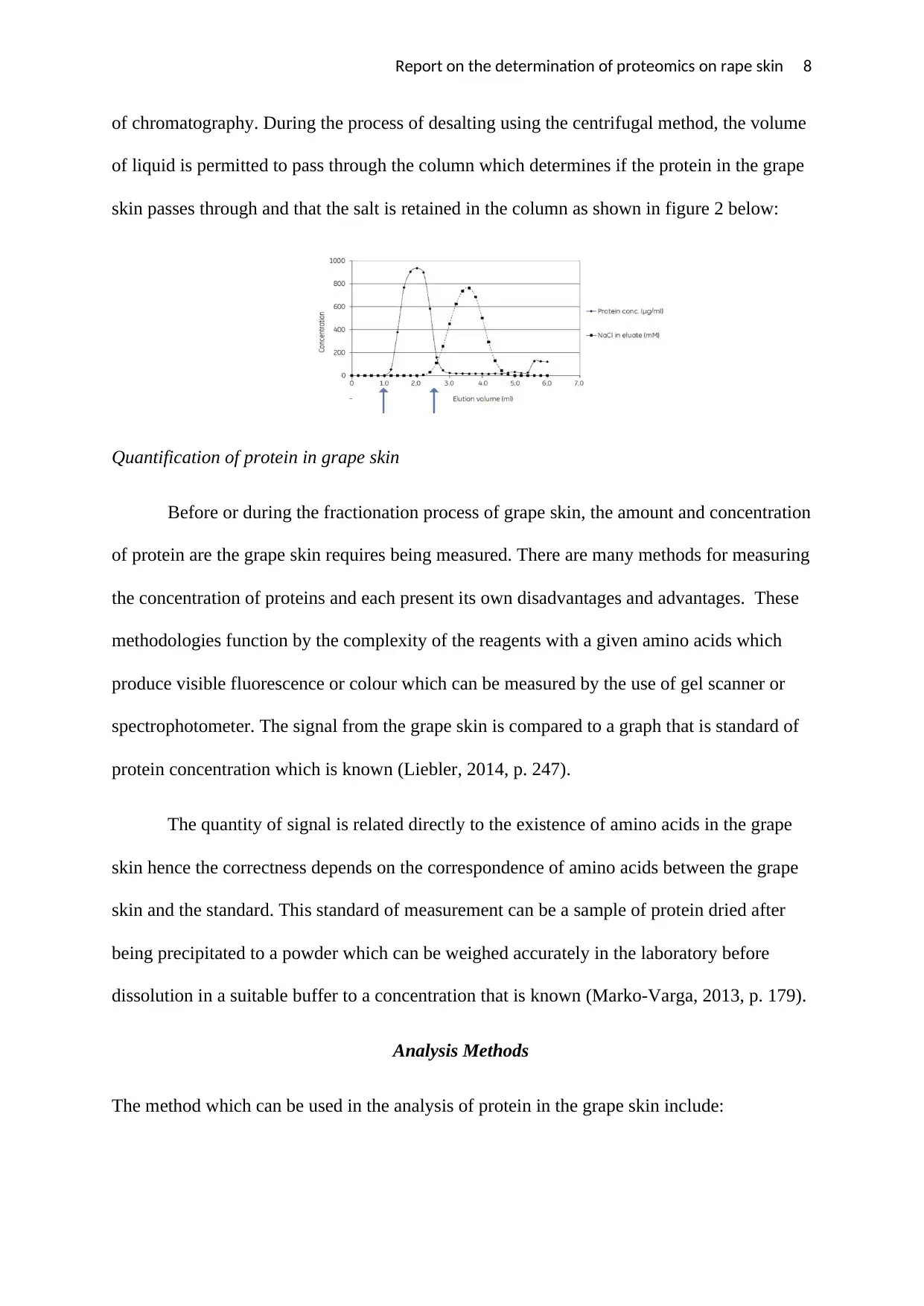
Report on the determination of proteomics on rape skin 8
of chromatography. During the process of desalting using the centrifugal method, the volume
of liquid is permitted to pass through the column which determines if the protein in the grape
skin passes through and that the salt is retained in the column as shown in figure 2 below:
Quantification of protein in grape skin
Before or during the fractionation process of grape skin, the amount and concentration
of protein are the grape skin requires being measured. There are many methods for measuring
the concentration of proteins and each present its own disadvantages and advantages. These
methodologies function by the complexity of the reagents with a given amino acids which
produce visible fluorescence or colour which can be measured by the use of gel scanner or
spectrophotometer. The signal from the grape skin is compared to a graph that is standard of
protein concentration which is known (Liebler, 2014, p. 247).
The quantity of signal is related directly to the existence of amino acids in the grape
skin hence the correctness depends on the correspondence of amino acids between the grape
skin and the standard. This standard of measurement can be a sample of protein dried after
being precipitated to a powder which can be weighed accurately in the laboratory before
dissolution in a suitable buffer to a concentration that is known (Marko-Varga, 2013, p. 179).
Analysis Methods
The method which can be used in the analysis of protein in the grape skin include:
of chromatography. During the process of desalting using the centrifugal method, the volume
of liquid is permitted to pass through the column which determines if the protein in the grape
skin passes through and that the salt is retained in the column as shown in figure 2 below:
Quantification of protein in grape skin
Before or during the fractionation process of grape skin, the amount and concentration
of protein are the grape skin requires being measured. There are many methods for measuring
the concentration of proteins and each present its own disadvantages and advantages. These
methodologies function by the complexity of the reagents with a given amino acids which
produce visible fluorescence or colour which can be measured by the use of gel scanner or
spectrophotometer. The signal from the grape skin is compared to a graph that is standard of
protein concentration which is known (Liebler, 2014, p. 247).
The quantity of signal is related directly to the existence of amino acids in the grape
skin hence the correctness depends on the correspondence of amino acids between the grape
skin and the standard. This standard of measurement can be a sample of protein dried after
being precipitated to a powder which can be weighed accurately in the laboratory before
dissolution in a suitable buffer to a concentration that is known (Marko-Varga, 2013, p. 179).
Analysis Methods
The method which can be used in the analysis of protein in the grape skin include:

Report on the determination of proteomics on rape skin 9
MS analysis: In this method, there is the use of the technique of chromatography which is
involved in the separation of peptide out before going through the mass spectrometer.
Comparative 2-DE: In this method, there is the separation of the proteins from the initial
sample which is complex.
SDS-PAGE: In this analysis method, the electrophoresis is used and have been proved to be
more effective because of the complexity of the skin. The SDS is used in making the protein
to be more soluble. During the process of separating the protein out, it is done by molecular
weight and not through points of isoelectric since the SDS wound affect it (Matthiesen, 2016,
p. 278).
The assays of proteins are sensitive to the reagents used in the preparation of the sample.
The alternative methods that can be used instead of this method are densitometry and 1D
PAGE in the determination of the concentration of the proteins. The methodology that has
been chosen in this report on proteomics of grape skin for measuring the proteins in 1D
PAGE due to some advantages which makes this method to be more effective and
advantageous that other methodologies of proteomics. These advantages include:
In this method, the sample can be excised, subjected to MS, and in-gel digested.
1D PAGE is companionable with numerous reagents used during the process of
preparation of the samples.
The outcome of the assay is comparable directly and comparative quantitation may
be done since 2D PAGE or 1D PAGE can be utilized during the process of
fractionation.
The gel fixing after the process of electrophoresis eliminates substance that is
contaminating which would affect the other assays (Michael Hamacher, 2014, p.
189).
SDS PAGE
MS analysis: In this method, there is the use of the technique of chromatography which is
involved in the separation of peptide out before going through the mass spectrometer.
Comparative 2-DE: In this method, there is the separation of the proteins from the initial
sample which is complex.
SDS-PAGE: In this analysis method, the electrophoresis is used and have been proved to be
more effective because of the complexity of the skin. The SDS is used in making the protein
to be more soluble. During the process of separating the protein out, it is done by molecular
weight and not through points of isoelectric since the SDS wound affect it (Matthiesen, 2016,
p. 278).
The assays of proteins are sensitive to the reagents used in the preparation of the sample.
The alternative methods that can be used instead of this method are densitometry and 1D
PAGE in the determination of the concentration of the proteins. The methodology that has
been chosen in this report on proteomics of grape skin for measuring the proteins in 1D
PAGE due to some advantages which makes this method to be more effective and
advantageous that other methodologies of proteomics. These advantages include:
In this method, the sample can be excised, subjected to MS, and in-gel digested.
1D PAGE is companionable with numerous reagents used during the process of
preparation of the samples.
The outcome of the assay is comparable directly and comparative quantitation may
be done since 2D PAGE or 1D PAGE can be utilized during the process of
fractionation.
The gel fixing after the process of electrophoresis eliminates substance that is
contaminating which would affect the other assays (Michael Hamacher, 2014, p.
189).
SDS PAGE
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Report on the determination of proteomics on rape skin 10
The methodology that has been chosen in this report on proteomics of grape skin for
measuring the proteins in 1D PAGE due to some advantages which makes this method to be
more effective and advantageous that other methodologies as stated above. The mass
spectrometer used in MS analysis has one great disadvantage which is that the ions being
measured by this instrument can only be measured one type at a given time. Some of the
provisions which the MS analysis have and other methodologies lack include the provision of
pH which makes the process of separation of proteins with other compounds hence efficient
separation (Misra, 2012, p. 179).
The commercially available mass spectrometers are also limited to certain ranges of
masses that is determined by a quadrupole. The MS analysis is advantageous than the other
two methodologies since it can be used to measure masses of both the proteins and peptides
hence providing complementary information of the peptides present in the grape skin as
opposed to the SDS PAGE which will only provide the mass of protein. The MS analysis also
has nanoelectrospray which is normally used in the analysis of mixtures of molecules which
are complex (Pedro Rodrigues, 2010, p. 278).
The SDS-PAGE which is an abbreviation of Sodium Dodecyl Sulfate-
PolyAcrylamide Gel Electrophoresis is the methodology suggested by this report of
measuring the mass of proteins in the grape skin. For the separation of the proteins by their
size, the molecules of the proteins should be coated with a compound which provides the
specific molecules with a similar overall charge so that the intrinsic charge of the proteins at a
given pH will not determine the process of separation.
Preparation of the grape skin sample
The grape skin should be boiled during the preparation of the sample for the SDS-
PAGE due to the urea presence since the proteins in the grape skin can denature at a
The methodology that has been chosen in this report on proteomics of grape skin for
measuring the proteins in 1D PAGE due to some advantages which makes this method to be
more effective and advantageous that other methodologies as stated above. The mass
spectrometer used in MS analysis has one great disadvantage which is that the ions being
measured by this instrument can only be measured one type at a given time. Some of the
provisions which the MS analysis have and other methodologies lack include the provision of
pH which makes the process of separation of proteins with other compounds hence efficient
separation (Misra, 2012, p. 179).
The commercially available mass spectrometers are also limited to certain ranges of
masses that is determined by a quadrupole. The MS analysis is advantageous than the other
two methodologies since it can be used to measure masses of both the proteins and peptides
hence providing complementary information of the peptides present in the grape skin as
opposed to the SDS PAGE which will only provide the mass of protein. The MS analysis also
has nanoelectrospray which is normally used in the analysis of mixtures of molecules which
are complex (Pedro Rodrigues, 2010, p. 278).
The SDS-PAGE which is an abbreviation of Sodium Dodecyl Sulfate-
PolyAcrylamide Gel Electrophoresis is the methodology suggested by this report of
measuring the mass of proteins in the grape skin. For the separation of the proteins by their
size, the molecules of the proteins should be coated with a compound which provides the
specific molecules with a similar overall charge so that the intrinsic charge of the proteins at a
given pH will not determine the process of separation.
Preparation of the grape skin sample
The grape skin should be boiled during the preparation of the sample for the SDS-
PAGE due to the urea presence since the proteins in the grape skin can denature at a
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Report on the determination of proteomics on rape skin 11
temperature of 20 degrees. The SDS is added in the compound so as to coat the proteins and
enabling their migration by the gel to be as a result of size alone. The digestion of proteomic
into minute peptides is important for the fractionation of the protein and for sensitivity,
detection, and measurement of the mass spectrometer (Sechi, 2011, p. 258).
There are a huge quantity of available enzymes of proteomic, however, a great
number of projects involving proteomic is done by the use of trypsin due to the following
properties: it is a minute protein hence can diffuse into gel, slight specificity which is defined,
and cutting at arginine and lysine produces peptides possessing group of amine at each end of
peptide. During the stage of isometric focusing by the use of pH gradient strip that is
immobilised, the initial consideration is the determination of the load of protein that is to be
used and the IPG gradient suitable for the grape skin being investigated which is shown in
table 2 below:
Type of stain Concentration of protein in mg/ml
Silver staining 0.1 to 1.5
SYPRO ruby 0.1 to 1.5
Coomassie Blue 0.5 to 2.5
The pH gradient and the IPG length depend on the features of the grape skin sample.
The trip of IPG requires to be rehydrated and there are options for which this method being
either passive or actively rehydration. During the stage of 2nd dimension SDS after IEF, the
major considerations in this stage are in relation to the choice of second gel dimension,
placement of the IPG strip, and equilibrium of the IPG strips (Sechi, 2011, p. 169).
Analysis by Mass Spectrometer and Peptide Fractionation
The mass spectrometer measure mass of ions or molecules which as charged. The ions
can be negatively or positively charged. The mass spectrometry is utilized in proteomics of
grape skin as a way of identification of what proteins are present in the grape skin which may
temperature of 20 degrees. The SDS is added in the compound so as to coat the proteins and
enabling their migration by the gel to be as a result of size alone. The digestion of proteomic
into minute peptides is important for the fractionation of the protein and for sensitivity,
detection, and measurement of the mass spectrometer (Sechi, 2011, p. 258).
There are a huge quantity of available enzymes of proteomic, however, a great
number of projects involving proteomic is done by the use of trypsin due to the following
properties: it is a minute protein hence can diffuse into gel, slight specificity which is defined,
and cutting at arginine and lysine produces peptides possessing group of amine at each end of
peptide. During the stage of isometric focusing by the use of pH gradient strip that is
immobilised, the initial consideration is the determination of the load of protein that is to be
used and the IPG gradient suitable for the grape skin being investigated which is shown in
table 2 below:
Type of stain Concentration of protein in mg/ml
Silver staining 0.1 to 1.5
SYPRO ruby 0.1 to 1.5
Coomassie Blue 0.5 to 2.5
The pH gradient and the IPG length depend on the features of the grape skin sample.
The trip of IPG requires to be rehydrated and there are options for which this method being
either passive or actively rehydration. During the stage of 2nd dimension SDS after IEF, the
major considerations in this stage are in relation to the choice of second gel dimension,
placement of the IPG strip, and equilibrium of the IPG strips (Sechi, 2011, p. 169).
Analysis by Mass Spectrometer and Peptide Fractionation
The mass spectrometer measure mass of ions or molecules which as charged. The ions
can be negatively or positively charged. The mass spectrometry is utilized in proteomics of
grape skin as a way of identification of what proteins are present in the grape skin which may
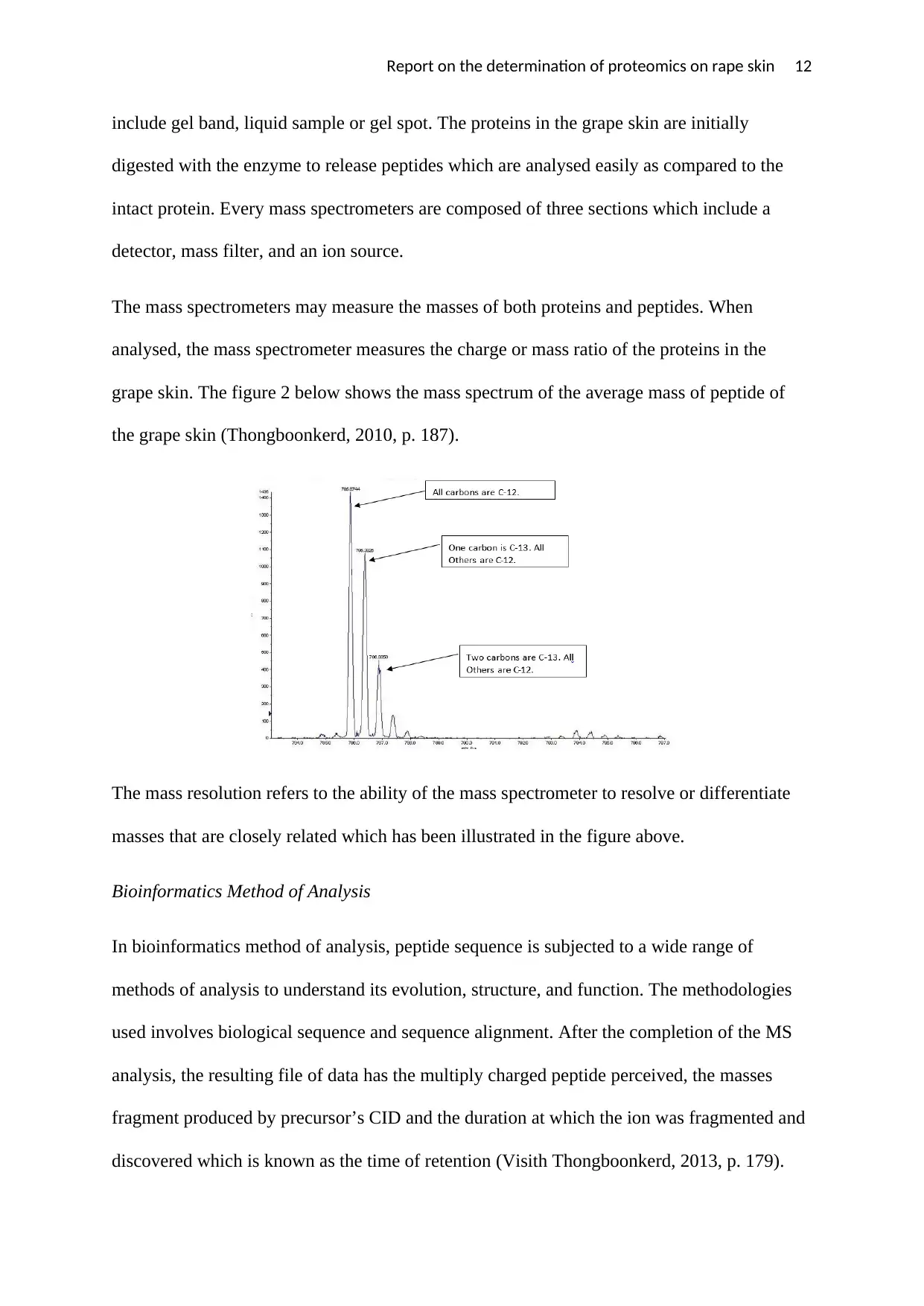
Report on the determination of proteomics on rape skin 12
include gel band, liquid sample or gel spot. The proteins in the grape skin are initially
digested with the enzyme to release peptides which are analysed easily as compared to the
intact protein. Every mass spectrometers are composed of three sections which include a
detector, mass filter, and an ion source.
The mass spectrometers may measure the masses of both proteins and peptides. When
analysed, the mass spectrometer measures the charge or mass ratio of the proteins in the
grape skin. The figure 2 below shows the mass spectrum of the average mass of peptide of
the grape skin (Thongboonkerd, 2010, p. 187).
The mass resolution refers to the ability of the mass spectrometer to resolve or differentiate
masses that are closely related which has been illustrated in the figure above.
Bioinformatics Method of Analysis
In bioinformatics method of analysis, peptide sequence is subjected to a wide range of
methods of analysis to understand its evolution, structure, and function. The methodologies
used involves biological sequence and sequence alignment. After the completion of the MS
analysis, the resulting file of data has the multiply charged peptide perceived, the masses
fragment produced by precursor’s CID and the duration at which the ion was fragmented and
discovered which is known as the time of retention (Visith Thongboonkerd, 2013, p. 179).
include gel band, liquid sample or gel spot. The proteins in the grape skin are initially
digested with the enzyme to release peptides which are analysed easily as compared to the
intact protein. Every mass spectrometers are composed of three sections which include a
detector, mass filter, and an ion source.
The mass spectrometers may measure the masses of both proteins and peptides. When
analysed, the mass spectrometer measures the charge or mass ratio of the proteins in the
grape skin. The figure 2 below shows the mass spectrum of the average mass of peptide of
the grape skin (Thongboonkerd, 2010, p. 187).
The mass resolution refers to the ability of the mass spectrometer to resolve or differentiate
masses that are closely related which has been illustrated in the figure above.
Bioinformatics Method of Analysis
In bioinformatics method of analysis, peptide sequence is subjected to a wide range of
methods of analysis to understand its evolution, structure, and function. The methodologies
used involves biological sequence and sequence alignment. After the completion of the MS
analysis, the resulting file of data has the multiply charged peptide perceived, the masses
fragment produced by precursor’s CID and the duration at which the ion was fragmented and
discovered which is known as the time of retention (Visith Thongboonkerd, 2013, p. 179).
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 15
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.





