Comparison of Methods for Detecting SNP (rs4680) in COMT Gene
VerifiedAdded on 2023/04/21
|27
|6924
|261
Report
AI Summary
This report presents an evaluation of various methods used to detect the single nucleotide polymorphism (SNP) rs4680 within the catechol-O-methyltransferase (COMT) gene. The study explores several techniques, including allele-specific PCR, high-resolution melt (HRM) analysis, restriction fragment length polymorphism (RFLP) analysis, Sanger sequencing, and TaqMan probe-based methods, to assess their efficiency in identifying the SNP. The report provides an overview of the COMT gene's function in degrading catecholamines, the significance of the rs4680 SNP, and its impact on the resultant enzyme's structure and activity. The analysis includes a comparison of the methods, considering factors such as cost, limitations, and suitability for laboratory and hospital settings, culminating in recommendations for the most appropriate techniques for routine SNP testing.

University
*** Semester
Genetics
Student Name:
Register Number:
Submission Date:
*** Semester
Genetics
Student Name:
Register Number:
Submission Date:
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Table of Contents
Introduction..................................................................................................................................................................... 3
Evaluation of Various Methods used to Detect SNP (rs4680).............................................................................................5
Abstract........................................................................................................................................................................ 5
Introduction................................................................................................................................................................. 5
Methods for Genotyping Single Nucleotide Polymorphisms...........................................................................................8
Materials and Methods................................................................................................................................................. 9
Result and Discussion................................................................................................................................................... 9
Comparison of the Five Methods in this Study............................................................................................................13
REFERENCES.............................................................................................................................................................. 16
Analysis of changes in E-cadherin and vimentin mRNA expression in human hepatocellular carcinoma cell...................18
Abstract...................................................................................................................................................................... 18
Introduction............................................................................................................................................................... 18
Materials and Methods............................................................................................................................................... 19
Results........................................................................................................................................................................ 19
Discussion................................................................................................................................................................... 24
Overall Conclusion......................................................................................................................................................... 26
REFERENCES.............................................................................................................................................................. 26
Introduction..................................................................................................................................................................... 3
Evaluation of Various Methods used to Detect SNP (rs4680).............................................................................................5
Abstract........................................................................................................................................................................ 5
Introduction................................................................................................................................................................. 5
Methods for Genotyping Single Nucleotide Polymorphisms...........................................................................................8
Materials and Methods................................................................................................................................................. 9
Result and Discussion................................................................................................................................................... 9
Comparison of the Five Methods in this Study............................................................................................................13
REFERENCES.............................................................................................................................................................. 16
Analysis of changes in E-cadherin and vimentin mRNA expression in human hepatocellular carcinoma cell...................18
Abstract...................................................................................................................................................................... 18
Introduction............................................................................................................................................................... 18
Materials and Methods............................................................................................................................................... 19
Results........................................................................................................................................................................ 19
Discussion................................................................................................................................................................... 24
Overall Conclusion......................................................................................................................................................... 26
REFERENCES.............................................................................................................................................................. 26

Introduction
In the field of medicine, the therapeutics, diagnostics and treatment of diseases have
advanced with the biomedical techniques. Moreover, the general research and translating the
cellular systems’ various mysteries with thorough investigation has advanced. In molecular
biology, the advent of new techniques has helped in analyzing the DNA and RNA structures.
Similarly, the other related methods allow to identify many parameters while testing. It can be
applied to screen the chromosome anomalies, mutations, differences or changes in the
functioning of protein, and mainly benefits the biomedical industry, forensics, genetics,
molecular biological research, evolution and others. This research ensures to find the analysis of
DNA and RNA. Hence, this technology is helpful for the crime scene investigation, where the
suspect’s DNA is utilized by the forensic team. In any case, it is a significant strategy utilized in
the investigation of genetics, DNA examination of various living beings, identification of protein
and recognition of ailments. PCR engages the utilization of methods to increase the measure of
DNA and subsequently decide distinguishing proof, and in addition being fundamental in atomic
hereditary examination (En.unifg.it, 2014).
The usually utilized atomic science methods are, Electrophoresis, PCR, Restriction
Digest, Ligation, Blotting and Cloning. This task researches various ordinarily utilized atomic
science systems including DNA and RNA. Polymerase Chain Reaction (PCR) – a procedure used
to enhance little measures of DNA to sums which can be utilized in further trials. It is utilized as
an essential device in atomic science to guarantee that adequate DNA is available to complete the
further strategies (Diamantina Institute, 2017).
Ribonucleic destructive or RNA is one of the three vital normal macromolecules that are
essential for each and every known kind of life. A central statute of sub-nuclear science
communicates that the flood of innate information in a cell is from DNA through RNA to
proteins: "DNA makes RNA makes protein". Proteins are the workhorses of the telephone; they
accept driving occupations in the telephone as mixes, as helper portions, and in cell motioning, to
give a few precedents.
DNA is considered as the "plot" of the cell; it passes on most of the genetic information
required for the cell to create, to take in enhancements, and to spread. RNA, in this role is the
"DNA photocopy" of the cell. Exactly when the cell needs to convey an explicit protein, it incites
the protein's gene the portion of DNA that codes for that protein– and produces distinctive copies
In the field of medicine, the therapeutics, diagnostics and treatment of diseases have
advanced with the biomedical techniques. Moreover, the general research and translating the
cellular systems’ various mysteries with thorough investigation has advanced. In molecular
biology, the advent of new techniques has helped in analyzing the DNA and RNA structures.
Similarly, the other related methods allow to identify many parameters while testing. It can be
applied to screen the chromosome anomalies, mutations, differences or changes in the
functioning of protein, and mainly benefits the biomedical industry, forensics, genetics,
molecular biological research, evolution and others. This research ensures to find the analysis of
DNA and RNA. Hence, this technology is helpful for the crime scene investigation, where the
suspect’s DNA is utilized by the forensic team. In any case, it is a significant strategy utilized in
the investigation of genetics, DNA examination of various living beings, identification of protein
and recognition of ailments. PCR engages the utilization of methods to increase the measure of
DNA and subsequently decide distinguishing proof, and in addition being fundamental in atomic
hereditary examination (En.unifg.it, 2014).
The usually utilized atomic science methods are, Electrophoresis, PCR, Restriction
Digest, Ligation, Blotting and Cloning. This task researches various ordinarily utilized atomic
science systems including DNA and RNA. Polymerase Chain Reaction (PCR) – a procedure used
to enhance little measures of DNA to sums which can be utilized in further trials. It is utilized as
an essential device in atomic science to guarantee that adequate DNA is available to complete the
further strategies (Diamantina Institute, 2017).
Ribonucleic destructive or RNA is one of the three vital normal macromolecules that are
essential for each and every known kind of life. A central statute of sub-nuclear science
communicates that the flood of innate information in a cell is from DNA through RNA to
proteins: "DNA makes RNA makes protein". Proteins are the workhorses of the telephone; they
accept driving occupations in the telephone as mixes, as helper portions, and in cell motioning, to
give a few precedents.
DNA is considered as the "plot" of the cell; it passes on most of the genetic information
required for the cell to create, to take in enhancements, and to spread. RNA, in this role is the
"DNA photocopy" of the cell. Exactly when the cell needs to convey an explicit protein, it incites
the protein's gene the portion of DNA that codes for that protein– and produces distinctive copies
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

of that bit of DNA as banner conveyor RNA, or mRNA (Rnasociety.org, 2019).
The primary lab incorporates, sanitization of the item with the assistance of TaqMan test
based frameworks, High Resolution Melt (HRM) examination, allele-explicit PCR, confinement
part length polymorphism (RFLP) investigation and sequencing were completed giving a strong
establishment and additionally the base for the utilization of the innovation in future applications.
The second lab incorporates the investigation of RNA. Thus, with RNA’s qPCR analysis by
using the gel electrophoresis, the cell culture, its methods and the RNA isolation process from
the cells and the preparation of cDNA synthesis were performed. Finally, the analysis of protein
is completed. This report aims to provide an updated theory and applications of various
laboratory techniques to identify the illness, where further investigation is carried out. After
SDS-PAGE and western blotting the comparative protein analysis between different cell lines
was conducted.
The essential stage in the lab course, included the disengagement of DNA, at that point
the utilization of the polymerase chain response (PCR) for DNA enhancement and gel
electrophoresis for recognizable proof of explicit proteins and their parts will be completed. Each
of these methods are utilized related to different procedures to enable the researchers to
illuminate some specific research questions (Diamantina Institute, 2017). Hence, these
technologies are extremely advanced and exposure to such techniques in the molecular genetic
level will help in the cellular machinery analysis. This can have effective applications such as to
detect the diseases, to identify protein malformations, for studying the mutation studies, genetics
and molecular biology.
The primary lab incorporates, sanitization of the item with the assistance of TaqMan test
based frameworks, High Resolution Melt (HRM) examination, allele-explicit PCR, confinement
part length polymorphism (RFLP) investigation and sequencing were completed giving a strong
establishment and additionally the base for the utilization of the innovation in future applications.
The second lab incorporates the investigation of RNA. Thus, with RNA’s qPCR analysis by
using the gel electrophoresis, the cell culture, its methods and the RNA isolation process from
the cells and the preparation of cDNA synthesis were performed. Finally, the analysis of protein
is completed. This report aims to provide an updated theory and applications of various
laboratory techniques to identify the illness, where further investigation is carried out. After
SDS-PAGE and western blotting the comparative protein analysis between different cell lines
was conducted.
The essential stage in the lab course, included the disengagement of DNA, at that point
the utilization of the polymerase chain response (PCR) for DNA enhancement and gel
electrophoresis for recognizable proof of explicit proteins and their parts will be completed. Each
of these methods are utilized related to different procedures to enable the researchers to
illuminate some specific research questions (Diamantina Institute, 2017). Hence, these
technologies are extremely advanced and exposure to such techniques in the molecular genetic
level will help in the cellular machinery analysis. This can have effective applications such as to
detect the diseases, to identify protein malformations, for studying the mutation studies, genetics
and molecular biology.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Evaluation of Various Methods used to Detect SNP (rs4680)
Abstract
Various enzymes along with catechol-O-methyltransferase (COMT) degrade
Catecholamines. In human beings COMT is using the COMT gene, which is present a couple of
isoforms namely, soluble short form (S-COMT) and long, membrane-bound form (MB-COMT).
In the COMT gene, rs4680 (Val158Met) refers to SNP. The COMT gene codes for the COMT
enzyme, breaks down the dopamine present in the brain's prefrontal cortex. The wild-type allele
is a (G), coding for a valine amino acid; the (A) substitution polymorphism changes the amino
acid to a methionine. Resulting in structure alteration of the resultant enzyme. Hence, the A allele
contains high dopamine in their prefrontal cortex that is responsible for various
neuropsychological associations (Infino.me, 2019). This report represents the experiments tested
in the lab and the reason for testing. The COMT is explained along with the relevance of SNP
rs4680. It determines how does the SNP results the change in behaviours. In the NCBI database
the COMT gene is identified along with the type of mutation, allele change and the location of
mutation on the gene. Necessary screenshots are represented for describing and showing the
understanding from the screenshot. It is observed how the SNP affect the protein. With the other
techniques, the control and gold standard results are compared, where the results are explained.
This study aims to examine the efficiency of allele-specific PCR, high resolution melt (HRM),
restriction fragment length polymorphism (RFLP), Sanger sequencing and TaqMan probe-based
methods, to detect rs4680 SNP in COMT.
Introduction
The mechanism of COMT in terms of degrading the catecholamines includes for
transferring a methyl group to it from the S-adenosyl methionine (SAM). In human beings
COMT is using the COMT gene, which is present a couple of isoforms namely, soluble short
form (S-COMT) and long, membrane-bound form (MB-COMT). COMT contains various
substrates, where all the compounds contain the structures of catechol like catecholestrogen and
flavonoids with the catechol moieties. In the NCBI database the COMT gene is identified along
with the type of mutation, allele change and the location of mutation on the gene.
The SNP stands for Single nuclear polymorphism, which are the common type of genetic
variation amongst people (Genetics Home Reference, 2019). The SNP detection technologies are
Abstract
Various enzymes along with catechol-O-methyltransferase (COMT) degrade
Catecholamines. In human beings COMT is using the COMT gene, which is present a couple of
isoforms namely, soluble short form (S-COMT) and long, membrane-bound form (MB-COMT).
In the COMT gene, rs4680 (Val158Met) refers to SNP. The COMT gene codes for the COMT
enzyme, breaks down the dopamine present in the brain's prefrontal cortex. The wild-type allele
is a (G), coding for a valine amino acid; the (A) substitution polymorphism changes the amino
acid to a methionine. Resulting in structure alteration of the resultant enzyme. Hence, the A allele
contains high dopamine in their prefrontal cortex that is responsible for various
neuropsychological associations (Infino.me, 2019). This report represents the experiments tested
in the lab and the reason for testing. The COMT is explained along with the relevance of SNP
rs4680. It determines how does the SNP results the change in behaviours. In the NCBI database
the COMT gene is identified along with the type of mutation, allele change and the location of
mutation on the gene. Necessary screenshots are represented for describing and showing the
understanding from the screenshot. It is observed how the SNP affect the protein. With the other
techniques, the control and gold standard results are compared, where the results are explained.
This study aims to examine the efficiency of allele-specific PCR, high resolution melt (HRM),
restriction fragment length polymorphism (RFLP), Sanger sequencing and TaqMan probe-based
methods, to detect rs4680 SNP in COMT.
Introduction
The mechanism of COMT in terms of degrading the catecholamines includes for
transferring a methyl group to it from the S-adenosyl methionine (SAM). In human beings
COMT is using the COMT gene, which is present a couple of isoforms namely, soluble short
form (S-COMT) and long, membrane-bound form (MB-COMT). COMT contains various
substrates, where all the compounds contain the structures of catechol like catecholestrogen and
flavonoids with the catechol moieties. In the NCBI database the COMT gene is identified along
with the type of mutation, allele change and the location of mutation on the gene.
The SNP stands for Single nuclear polymorphism, which are the common type of genetic
variation amongst people (Genetics Home Reference, 2019). The SNP detection technologies are

utilized for scanning new polymorphisms and for determining the allele(s) of a well-known
polymorphism in the target sequences. The SNP detection technologies are advanced based on
the labor, expensive processes and time consumed for the extremely automated, efficient, and
relatively inexpensive methods. SNP is a variation of a single base pair in the gene. The best
suitable example for SNP includes gene encoding catechol-O-methyltransferase (COMT), where
it is an enzyme which breaks down the neurotransmitters in the human brain. The SNP, rs4680, is
a single nucleotide variation, and it is found in COMT gene on the chromosome 22, whose
position is 19963748. The figure 1 represents this.
In COMT, Polymorphism rs4680 (Val158Met) arises from guanine’s (G) single base pair
substitution for adenine (A), which results in coding the methionine amino acid at the 158th
position of the genetic code instead of valine (Snpedia.com, 2019). Such a SNP modified the
COMT structure, which actually results in allele A (Met) that has reduced its activity of
functional enzyme when compared with its G allele’s COMT activity. With the transfer of methyl
group from an adenosyl methionine the COMT functions for the catecholamines which are
mainly found in the nervous system. Thus, COMT could be referred as the essential enzyme for
the body degradation. It works in terms of the catechol drug’s metabolism that is utilized for
treating the diseases like Parkinson. In a human body it is present in the following forms namely,
membrane-bound form and soluble form i.e., present in the circulation.
Here, the results will help to determine which assay works best for the laboratory and the
hospital, and an appropriate reason will be justified. Moving further, the cost is an effective
criteria to be known, so the cost of the processes will be identified, as the cost will help to know
which has best value for money while delivering the best results. The hospital’s criterions will be
addressed. Further, the techniques’ limitations are listed, including the solution to overcome the
issues. At last, recommendations for the hospital and lab are provided, related to the technique to
be utilized in the lab. From the below represented figure q, the A and G explicit alleles are
demonstrated with bioinformatics. The figure 2 and 3, represent the A and G specific alleles of
bioinformatics.
polymorphism in the target sequences. The SNP detection technologies are advanced based on
the labor, expensive processes and time consumed for the extremely automated, efficient, and
relatively inexpensive methods. SNP is a variation of a single base pair in the gene. The best
suitable example for SNP includes gene encoding catechol-O-methyltransferase (COMT), where
it is an enzyme which breaks down the neurotransmitters in the human brain. The SNP, rs4680, is
a single nucleotide variation, and it is found in COMT gene on the chromosome 22, whose
position is 19963748. The figure 1 represents this.
In COMT, Polymorphism rs4680 (Val158Met) arises from guanine’s (G) single base pair
substitution for adenine (A), which results in coding the methionine amino acid at the 158th
position of the genetic code instead of valine (Snpedia.com, 2019). Such a SNP modified the
COMT structure, which actually results in allele A (Met) that has reduced its activity of
functional enzyme when compared with its G allele’s COMT activity. With the transfer of methyl
group from an adenosyl methionine the COMT functions for the catecholamines which are
mainly found in the nervous system. Thus, COMT could be referred as the essential enzyme for
the body degradation. It works in terms of the catechol drug’s metabolism that is utilized for
treating the diseases like Parkinson. In a human body it is present in the following forms namely,
membrane-bound form and soluble form i.e., present in the circulation.
Here, the results will help to determine which assay works best for the laboratory and the
hospital, and an appropriate reason will be justified. Moving further, the cost is an effective
criteria to be known, so the cost of the processes will be identified, as the cost will help to know
which has best value for money while delivering the best results. The hospital’s criterions will be
addressed. Further, the techniques’ limitations are listed, including the solution to overcome the
issues. At last, recommendations for the hospital and lab are provided, related to the technique to
be utilized in the lab. From the below represented figure q, the A and G explicit alleles are
demonstrated with bioinformatics. The figure 2 and 3, represent the A and G specific alleles of
bioinformatics.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
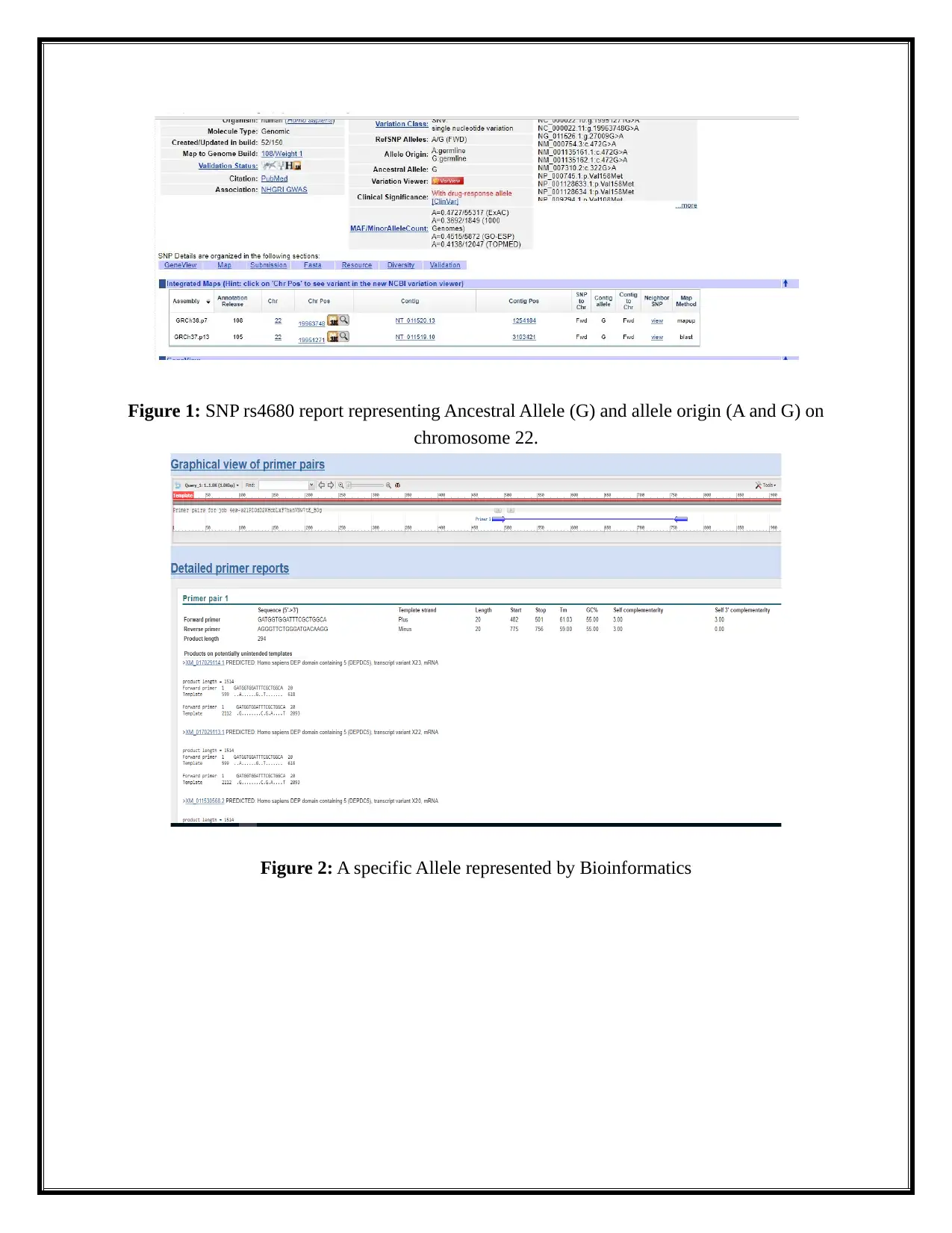
Figure 1: SNP rs4680 report representing Ancestral Allele (G) and allele origin (A and G) on
chromosome 22.
Figure 2: A specific Allele represented by Bioinformatics
chromosome 22.
Figure 2: A specific Allele represented by Bioinformatics
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Figure 3: G Specific Allele represented by Bioinformatics
Methods for Genotyping Single Nucleotide Polymorphisms
Genotyping refers to a technology which helps in the detection of small genetic
differences that could cause major changes in phenotype, along with both the physical
differences which make it stand out from other and represents the pathological changes
underlying the disease. Genotyping contains a large number of usages in the field of medicine,
agriculture and scientific research (Thermofisher.com, 2019). Based on the Human Genome
Project, these technologies are advancing. The almost completed human genome sequence gives
the reference against which all the other sequencing data could be compared. Based on either the
direct DNA sequencing or the denaturing high performance liquid chromatography (dHPLC), the
local, target, SNP discovery will be relied. Various SNP genotyping methods have evolved. The
SNP genotyping has high demand.
Allele-Specific PCR
Allele-specific PCR is a technique which uses the principle of Taq polymerases that needs the
hybridised primers at its 3’ end to work. It is utilized as the method of mutation detection, in
cases where the mutation site is known. It is helpful in establishing all the possible allele-specific
Methods for Genotyping Single Nucleotide Polymorphisms
Genotyping refers to a technology which helps in the detection of small genetic
differences that could cause major changes in phenotype, along with both the physical
differences which make it stand out from other and represents the pathological changes
underlying the disease. Genotyping contains a large number of usages in the field of medicine,
agriculture and scientific research (Thermofisher.com, 2019). Based on the Human Genome
Project, these technologies are advancing. The almost completed human genome sequence gives
the reference against which all the other sequencing data could be compared. Based on either the
direct DNA sequencing or the denaturing high performance liquid chromatography (dHPLC), the
local, target, SNP discovery will be relied. Various SNP genotyping methods have evolved. The
SNP genotyping has high demand.
Allele-Specific PCR
Allele-specific PCR is a technique which uses the principle of Taq polymerases that needs the
hybridised primers at its 3’ end to work. It is utilized as the method of mutation detection, in
cases where the mutation site is known. It is helpful in establishing all the possible allele-specific

amplification and genotypes (M et al., 2009).
Restriction Fragment Length Polymorphism (RFLP) Analysis
The analysis of RFLP refers to recognizing various homologous fragments based on the lengths,
post DNA digestion with the help of endonucleases.). In general, it analyzes the DNA fragments’
unique patterns for providing the genetically differentiation among different organisms. Such
unique patterns are known as Variable Number of Tandem Repeats (VNTRs) (Cheriyedath,
2018).
High Resolution Melt (HRM) Analysis
High Resolution Melt analysis implies the effective technique which is used to polymorphisms,
epigenetic and detect the mutations with the differences of double stranded DNA samples. HRM
contains large benefits when compared to the other genotyping technologies (Beginners Guide to
High Resolution Melt (HRM) analysis, n.d.).
TaqMan Probe Analysis
TaqMan probes are labeled as VIC and TAMRA on the 5′ nucleotide and 3′ nucleotide,
respectively (Sciencedirect.com, 2019). It is utilized for detecting the increase of fluorescence,
due to the dye’s release.
Sanger Sequencing
Sanger Sequencing is a method which revolves around the usage of polyacrylamide gels, for
releasing the products of DNA polymerase activity, which results in maximizing the length of the
chain (Valencia et al., 2013).
Materials and Methods
The Coventry University’s materials and methods are used.
Result and Discussion
A lab forms the patient's DNA tests sent from a close-by clinic. The healing center needs
to present testing for an explicit SNP (rs 4680) in the quality COMT anyway there are different
ways this can be cultivated. The line chief has asked for me and my associates to build up a
Restriction Fragment Length Polymorphism (RFLP) Analysis
The analysis of RFLP refers to recognizing various homologous fragments based on the lengths,
post DNA digestion with the help of endonucleases.). In general, it analyzes the DNA fragments’
unique patterns for providing the genetically differentiation among different organisms. Such
unique patterns are known as Variable Number of Tandem Repeats (VNTRs) (Cheriyedath,
2018).
High Resolution Melt (HRM) Analysis
High Resolution Melt analysis implies the effective technique which is used to polymorphisms,
epigenetic and detect the mutations with the differences of double stranded DNA samples. HRM
contains large benefits when compared to the other genotyping technologies (Beginners Guide to
High Resolution Melt (HRM) analysis, n.d.).
TaqMan Probe Analysis
TaqMan probes are labeled as VIC and TAMRA on the 5′ nucleotide and 3′ nucleotide,
respectively (Sciencedirect.com, 2019). It is utilized for detecting the increase of fluorescence,
due to the dye’s release.
Sanger Sequencing
Sanger Sequencing is a method which revolves around the usage of polyacrylamide gels, for
releasing the products of DNA polymerase activity, which results in maximizing the length of the
chain (Valencia et al., 2013).
Materials and Methods
The Coventry University’s materials and methods are used.
Result and Discussion
A lab forms the patient's DNA tests sent from a close-by clinic. The healing center needs
to present testing for an explicit SNP (rs 4680) in the quality COMT anyway there are different
ways this can be cultivated. The line chief has asked for me and my associates to build up a
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

measure to have the capacity of the routinely test for this SNP. However, the line supervisor and
doctor's facility has given me certain arrangement of criteria to pursue while building up this
analysis.
They need you to ponder which test works best for the research center and the doctor's
facility, at that point finish an answer to give your recommendations on which examine is ideal
and why.
A COMT inhibitor is a medication that hinders the activity of catechol-O-methyl. The
COMT quality gives directions to making a protein called catechol-O-methyltransferase. Two
forms of this compound are produced using the quality. The more drawn out frame, called film
bound catechol-O-methyltransferase (MB-COMT), is predominantly delivered by nerve cells in
the mind. Different tissues, including the liver, kidneys, and blood, create a shorter type of the
catalyst called dissolvable catechol-O-methyltransferase (S-COMT). This type of the protein
helps control the dimensions of specific hormones.
In the cerebrum, catechol-O-methyltransferase helps separate certain concoction delivery
people called synapses. These synthetic substances lead signals starting with one nerve cell then
onto the next. Catechol-O-methyltransferase is especially essential in a territory at the front of
the mind called the prefrontal cortex, which sorts out and organizes data from different parts of
the cerebrum. This district is included with identity, arranging, restraint of practices, dynamic
reasoning, feeling, and working (present moment) memory. To work proficiently, the prefrontal
cortex requires motioning by synapses, for example, dopamine and norepinephrine. Catechol-O-
methyltransferase keeps up suitable dimensions of these synapses in this piece of the brain
(Genetics Home Reference, 2019).
rs4680 (Val158Met) is a very much examined SNP in the COMT quality. The COMT
quality codes for the COMT catalyst, which separates dopamine in the mind's prefrontal cortex.
The wild-type allele is a (G), coding for a valine amino corrosive; the (A) substitution
polymorphism changes the amino corrosive to a methionine. This changes the structure of the
resultant catalyst with the end goal that its movement is just 25% of the wild sort. Thus, A allele
bearers have more dopamine in their prefrontal cortex, which might be in charge of huge
numbers of the neuropsychological affiliations recorded underneath.
23andMe blog condenses the alleles at this SNP as (Snpedia.com, 2019),
rs4680(A) = Worrier. Met, increasingly exploratory, bring down COMT enzymatic action,
doctor's facility has given me certain arrangement of criteria to pursue while building up this
analysis.
They need you to ponder which test works best for the research center and the doctor's
facility, at that point finish an answer to give your recommendations on which examine is ideal
and why.
A COMT inhibitor is a medication that hinders the activity of catechol-O-methyl. The
COMT quality gives directions to making a protein called catechol-O-methyltransferase. Two
forms of this compound are produced using the quality. The more drawn out frame, called film
bound catechol-O-methyltransferase (MB-COMT), is predominantly delivered by nerve cells in
the mind. Different tissues, including the liver, kidneys, and blood, create a shorter type of the
catalyst called dissolvable catechol-O-methyltransferase (S-COMT). This type of the protein
helps control the dimensions of specific hormones.
In the cerebrum, catechol-O-methyltransferase helps separate certain concoction delivery
people called synapses. These synthetic substances lead signals starting with one nerve cell then
onto the next. Catechol-O-methyltransferase is especially essential in a territory at the front of
the mind called the prefrontal cortex, which sorts out and organizes data from different parts of
the cerebrum. This district is included with identity, arranging, restraint of practices, dynamic
reasoning, feeling, and working (present moment) memory. To work proficiently, the prefrontal
cortex requires motioning by synapses, for example, dopamine and norepinephrine. Catechol-O-
methyltransferase keeps up suitable dimensions of these synapses in this piece of the brain
(Genetics Home Reference, 2019).
rs4680 (Val158Met) is a very much examined SNP in the COMT quality. The COMT
quality codes for the COMT catalyst, which separates dopamine in the mind's prefrontal cortex.
The wild-type allele is a (G), coding for a valine amino corrosive; the (A) substitution
polymorphism changes the amino corrosive to a methionine. This changes the structure of the
resultant catalyst with the end goal that its movement is just 25% of the wild sort. Thus, A allele
bearers have more dopamine in their prefrontal cortex, which might be in charge of huge
numbers of the neuropsychological affiliations recorded underneath.
23andMe blog condenses the alleles at this SNP as (Snpedia.com, 2019),
rs4680(A) = Worrier. Met, increasingly exploratory, bring down COMT enzymatic action,
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

along these lines higher dopamine levels; bring down torment limit, upgraded
helplessness to push, yet additionally progressively proficient at preparing data under
generally conditions
rs4680(G) = Warrior. Val, less exploratory, higher COMT enzymatic action, subsequently
bring down dopamine levels; higher torment edge, better pressure strength, but with a
humble decrease in official cognizance execution under generally conditions
A SNP is an adjustment in 1 nucleotide or base-combine inside a codon in the DNA (Akey et al.,
2001). Contingent upon its area, a SNP may adjust how a quality is deciphered or the amino
corrosive succession for the protein being made, at last causing an adjustment in action of that
protein. Sadly, SNPs can happen with numerous proteins associated with medication transport,
digestion and receptors that at last impact both the pharmacokinetic and pharmacodynamic
properties of numerous medications (Ebmconsult.com, 2019). It is assumed that the control
samples might have produced three types of genotypes i.e., AA, AG, GG. But, for the controls,
generation of consistent results by the five methods actually failed. This implies that the samples
could have been contaminated. However, AG and GG genotypes were produced by Sanger
method.
The table 1, tabulates the results of the five SNP genotyping methods which are applied
on nine tests and three control samples, for identifying rs4680 SNP of COMT. The different
genotype methods represented in the below table are listed below:
1) Restriction Fragment Length Polymorphism (RFLP) Analysis
2) Allele-Specific PCR
3) High Resolution Melt (HRM) Analysis
4) TaqMan Probe Analysis
5) Sanger Sequencing
It is observed that the results were same for AE, RAP, DEP, SH, VA and TJ where just
four methods were able to effectively identify SNP. Only for GG homozygous genotype, DEP
and RAP found the result with 4 assay methods, but with the help AS method no results were
found. Just the AG heterozygous genotypes were recognized from the four methods which
generated the results for VA and TG samples. Further, it is observed that for these samples the
helplessness to push, yet additionally progressively proficient at preparing data under
generally conditions
rs4680(G) = Warrior. Val, less exploratory, higher COMT enzymatic action, subsequently
bring down dopamine levels; higher torment edge, better pressure strength, but with a
humble decrease in official cognizance execution under generally conditions
A SNP is an adjustment in 1 nucleotide or base-combine inside a codon in the DNA (Akey et al.,
2001). Contingent upon its area, a SNP may adjust how a quality is deciphered or the amino
corrosive succession for the protein being made, at last causing an adjustment in action of that
protein. Sadly, SNPs can happen with numerous proteins associated with medication transport,
digestion and receptors that at last impact both the pharmacokinetic and pharmacodynamic
properties of numerous medications (Ebmconsult.com, 2019). It is assumed that the control
samples might have produced three types of genotypes i.e., AA, AG, GG. But, for the controls,
generation of consistent results by the five methods actually failed. This implies that the samples
could have been contaminated. However, AG and GG genotypes were produced by Sanger
method.
The table 1, tabulates the results of the five SNP genotyping methods which are applied
on nine tests and three control samples, for identifying rs4680 SNP of COMT. The different
genotype methods represented in the below table are listed below:
1) Restriction Fragment Length Polymorphism (RFLP) Analysis
2) Allele-Specific PCR
3) High Resolution Melt (HRM) Analysis
4) TaqMan Probe Analysis
5) Sanger Sequencing
It is observed that the results were same for AE, RAP, DEP, SH, VA and TJ where just
four methods were able to effectively identify SNP. Only for GG homozygous genotype, DEP
and RAP found the result with 4 assay methods, but with the help AS method no results were
found. Just the AG heterozygous genotypes were recognized from the four methods which
generated the results for VA and TG samples. Further, it is observed that for these samples the
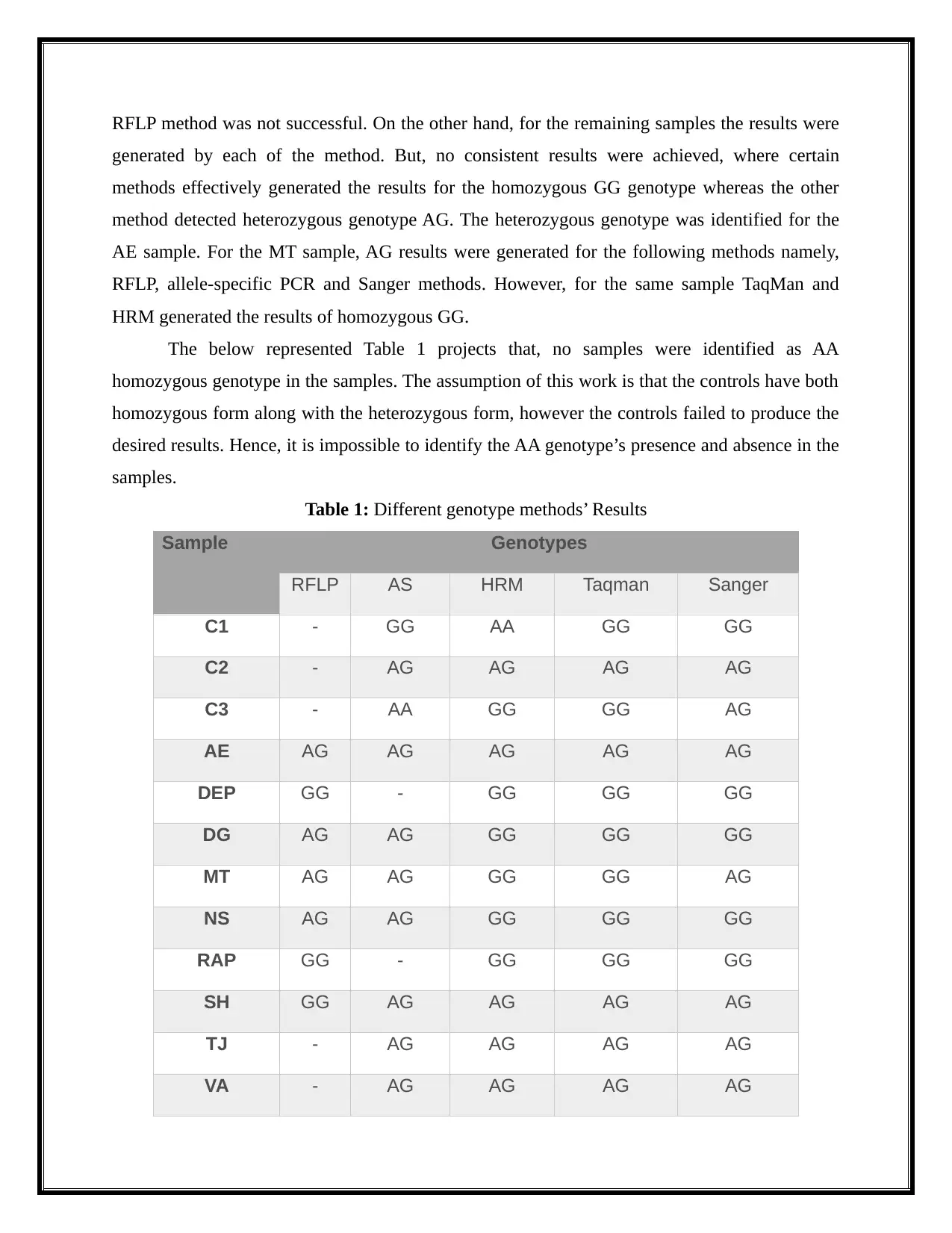
RFLP method was not successful. On the other hand, for the remaining samples the results were
generated by each of the method. But, no consistent results were achieved, where certain
methods effectively generated the results for the homozygous GG genotype whereas the other
method detected heterozygous genotype AG. The heterozygous genotype was identified for the
AE sample. For the MT sample, AG results were generated for the following methods namely,
RFLP, allele-specific PCR and Sanger methods. However, for the same sample TaqMan and
HRM generated the results of homozygous GG.
The below represented Table 1 projects that, no samples were identified as AA
homozygous genotype in the samples. The assumption of this work is that the controls have both
homozygous form along with the heterozygous form, however the controls failed to produce the
desired results. Hence, it is impossible to identify the AA genotype’s presence and absence in the
samples.
Table 1: Different genotype methods’ Results
Sample Genotypes
RFLP AS HRM Taqman Sanger
C1 - GG AA GG GG
C2 - AG AG AG AG
C3 - AA GG GG AG
AE AG AG AG AG AG
DEP GG - GG GG GG
DG AG AG GG GG GG
MT AG AG GG GG AG
NS AG AG GG GG GG
RAP GG - GG GG GG
SH GG AG AG AG AG
TJ - AG AG AG AG
VA - AG AG AG AG
generated by each of the method. But, no consistent results were achieved, where certain
methods effectively generated the results for the homozygous GG genotype whereas the other
method detected heterozygous genotype AG. The heterozygous genotype was identified for the
AE sample. For the MT sample, AG results were generated for the following methods namely,
RFLP, allele-specific PCR and Sanger methods. However, for the same sample TaqMan and
HRM generated the results of homozygous GG.
The below represented Table 1 projects that, no samples were identified as AA
homozygous genotype in the samples. The assumption of this work is that the controls have both
homozygous form along with the heterozygous form, however the controls failed to produce the
desired results. Hence, it is impossible to identify the AA genotype’s presence and absence in the
samples.
Table 1: Different genotype methods’ Results
Sample Genotypes
RFLP AS HRM Taqman Sanger
C1 - GG AA GG GG
C2 - AG AG AG AG
C3 - AA GG GG AG
AE AG AG AG AG AG
DEP GG - GG GG GG
DG AG AG GG GG GG
MT AG AG GG GG AG
NS AG AG GG GG GG
RAP GG - GG GG GG
SH GG AG AG AG AG
TJ - AG AG AG AG
VA - AG AG AG AG
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 27
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.




