Detailed Comparison of Eukaryotic and Prokaryotic Gene Expression
VerifiedAdded on 2023/05/28
|23
|1356
|154
Essay
AI Summary
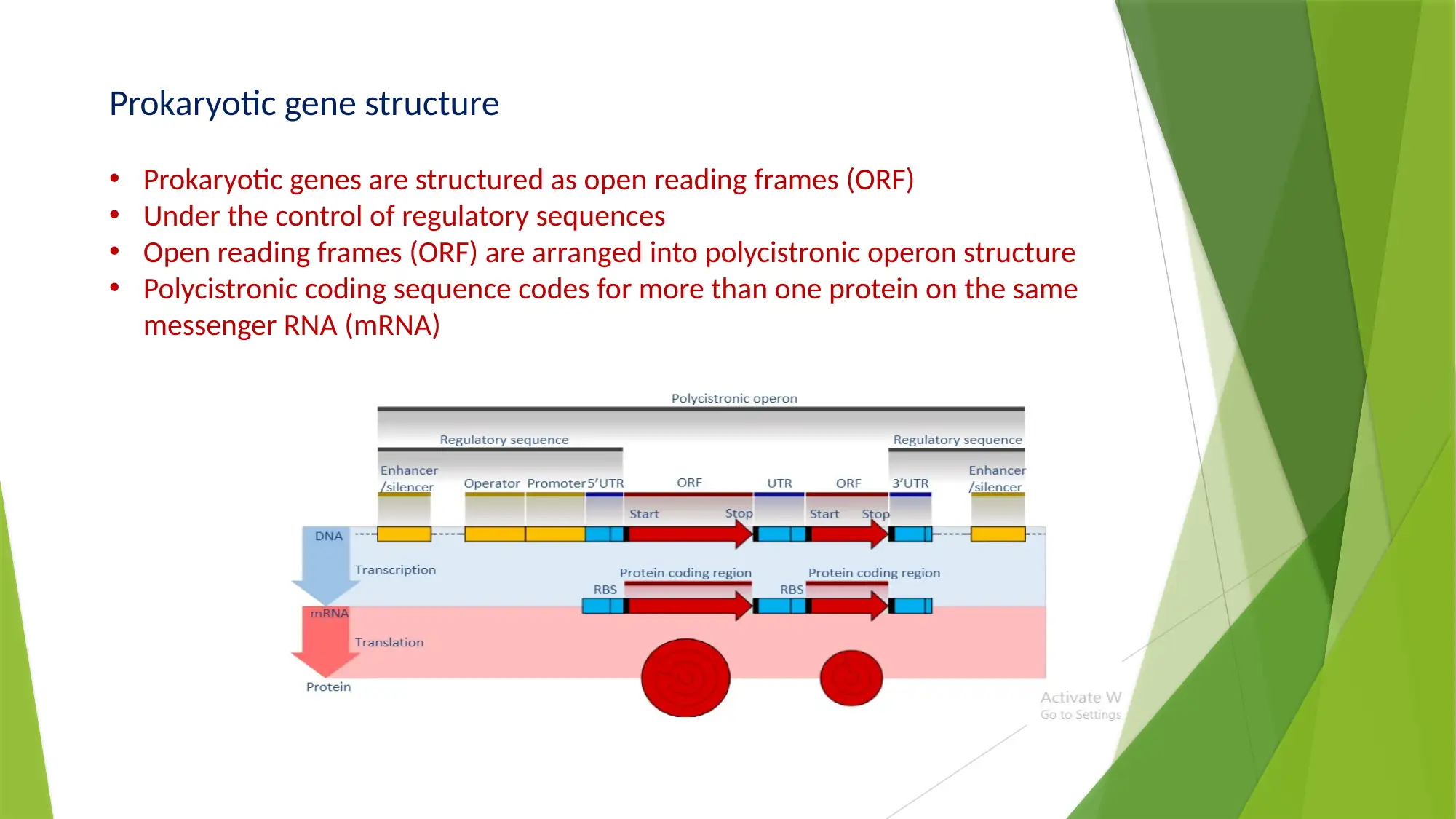
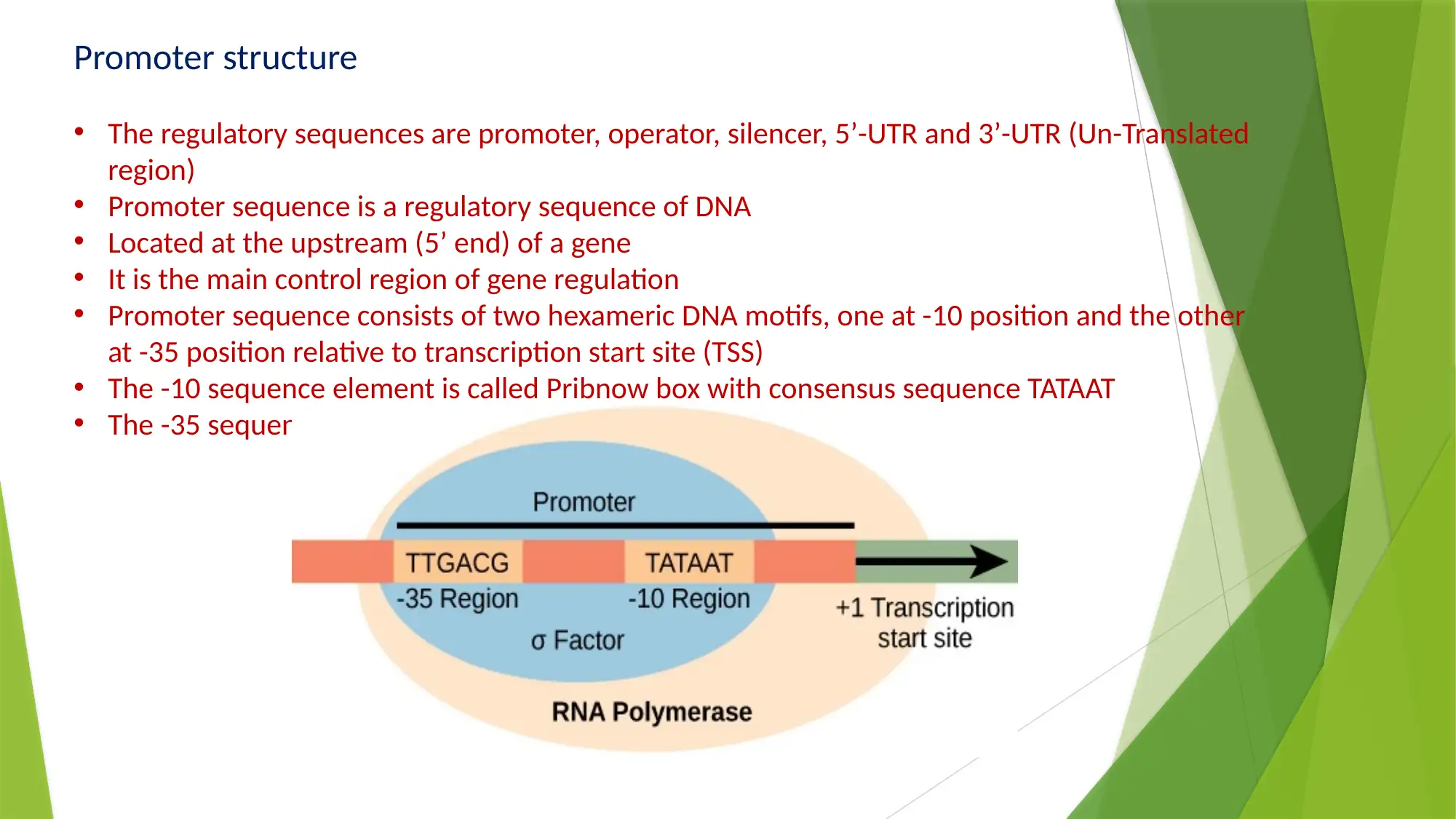
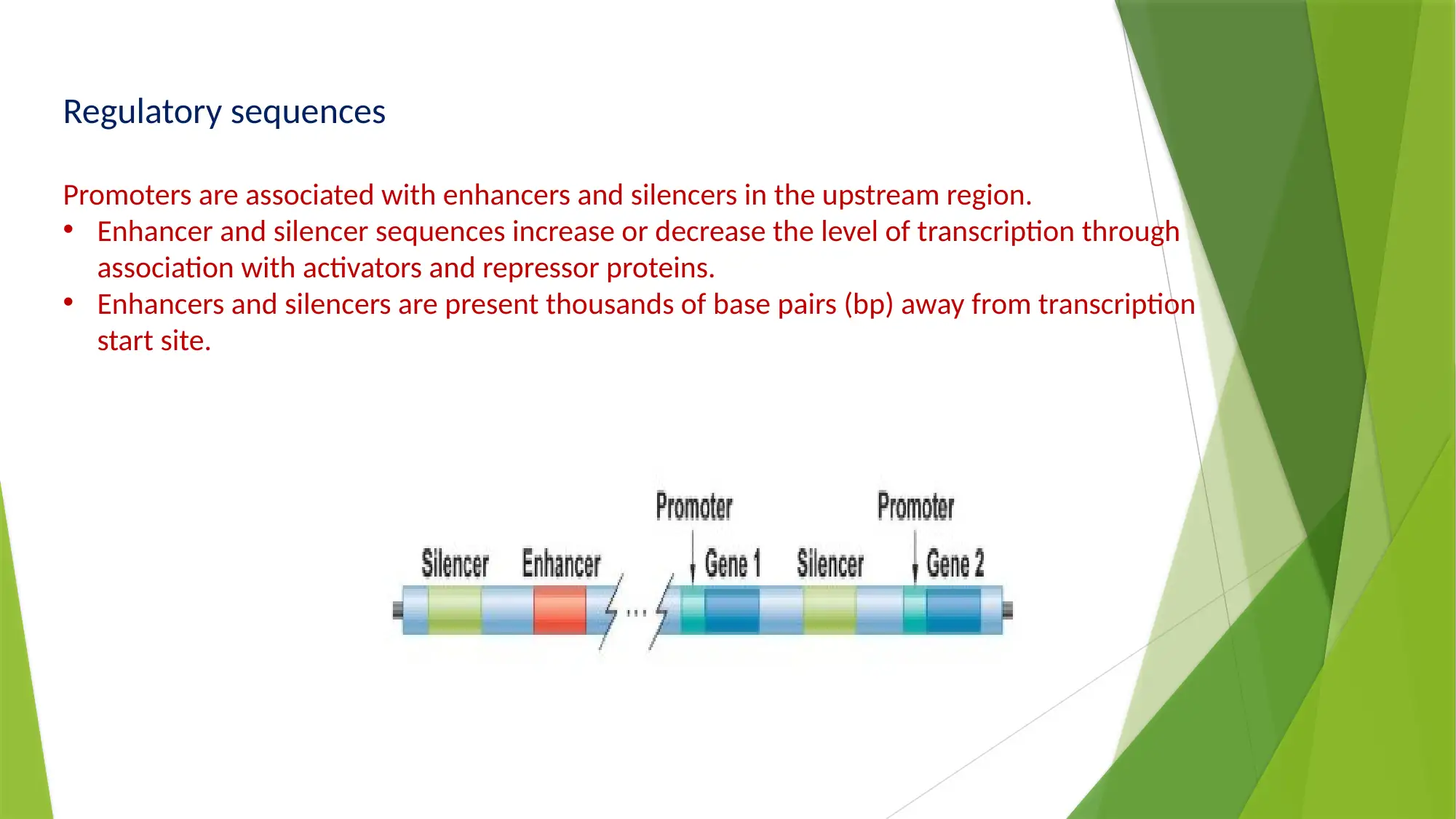
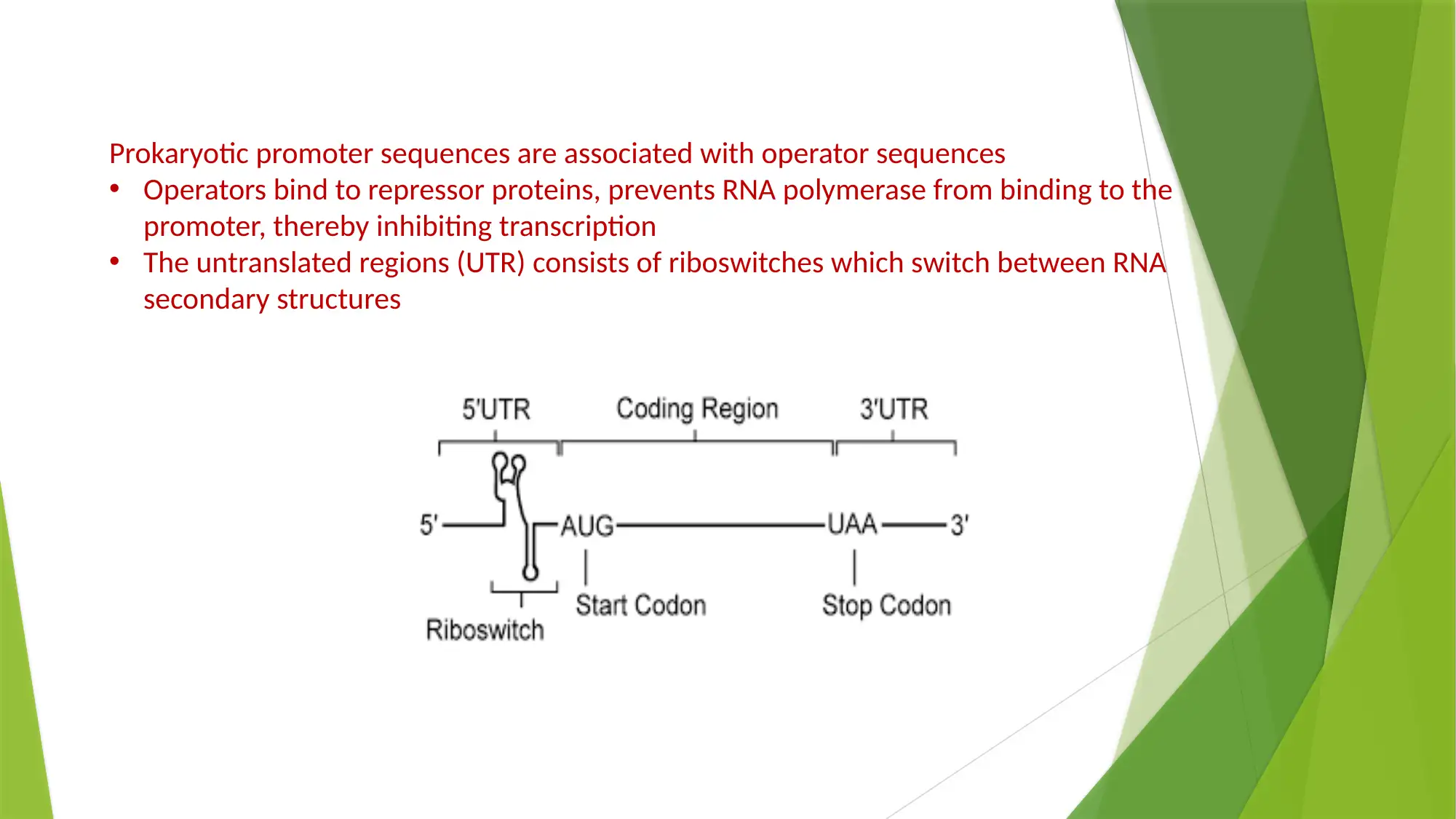
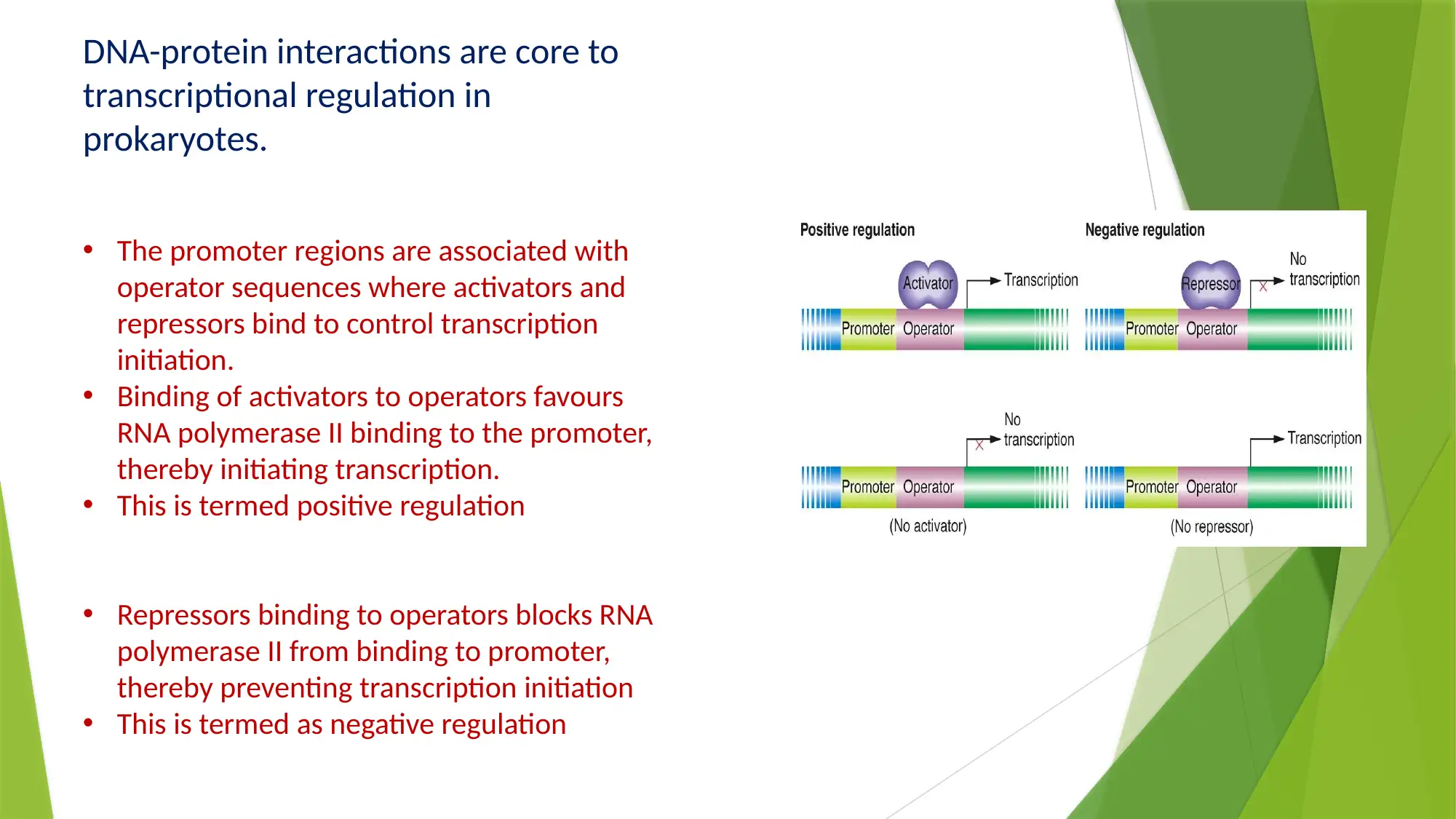
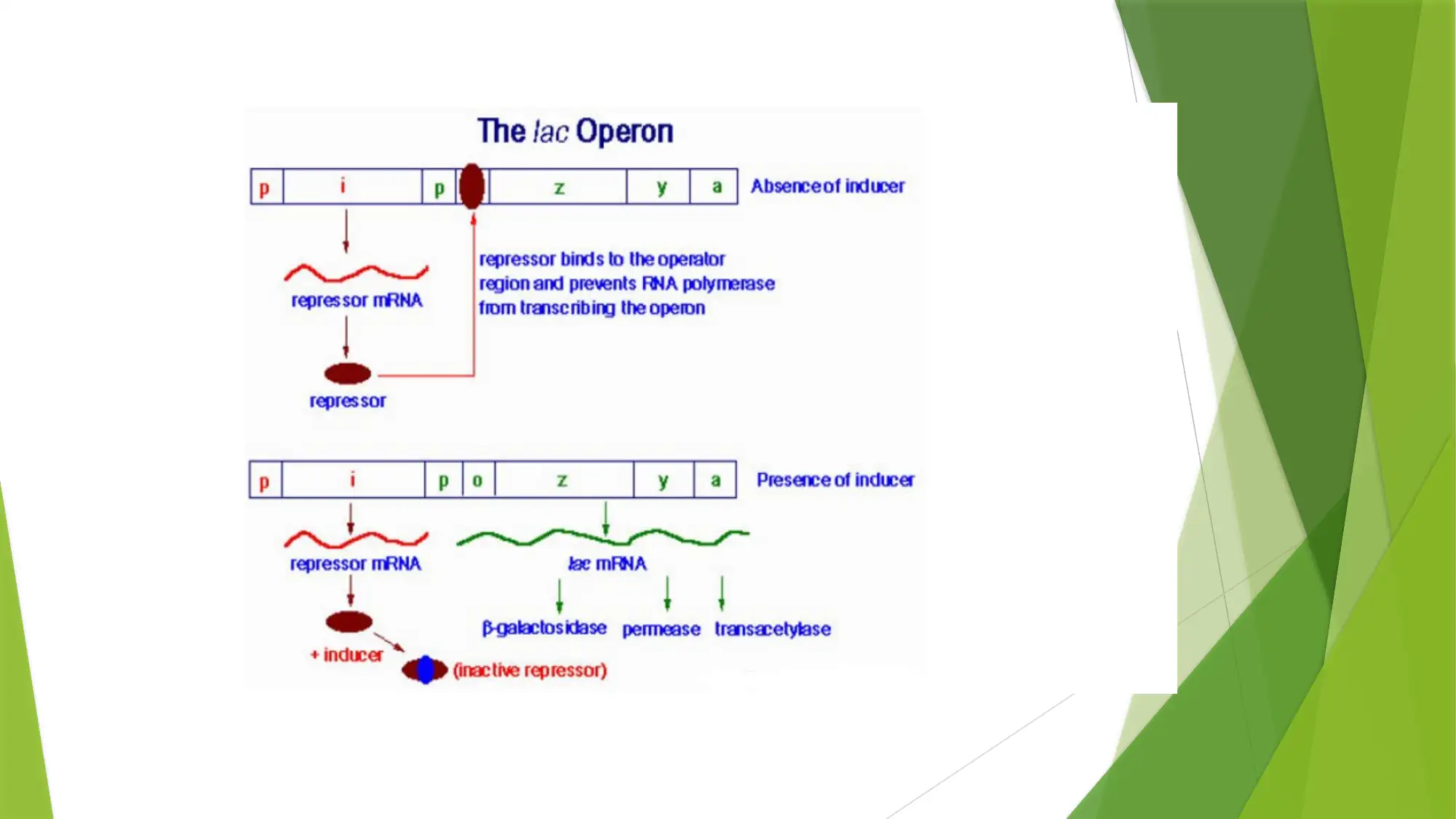
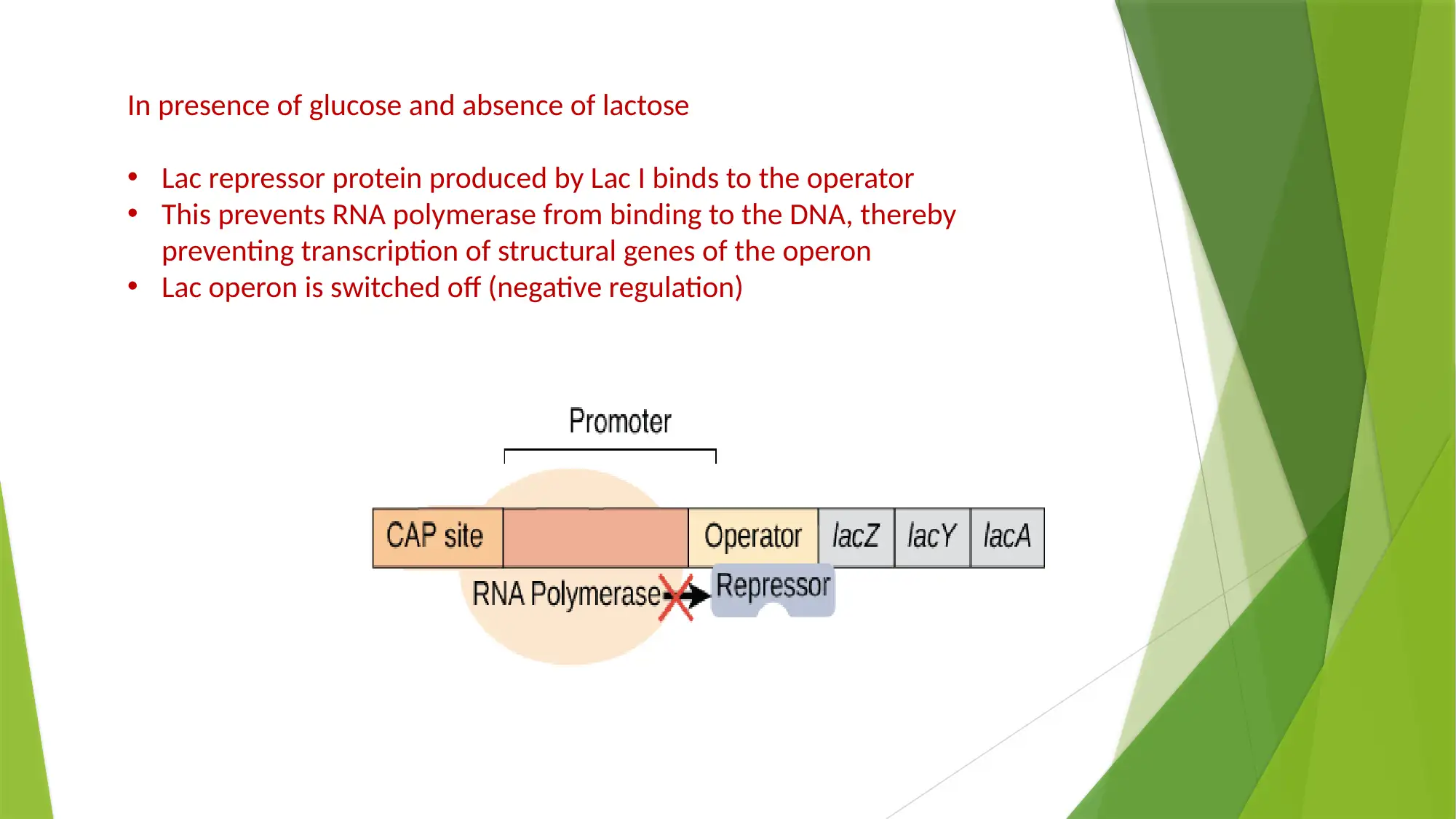
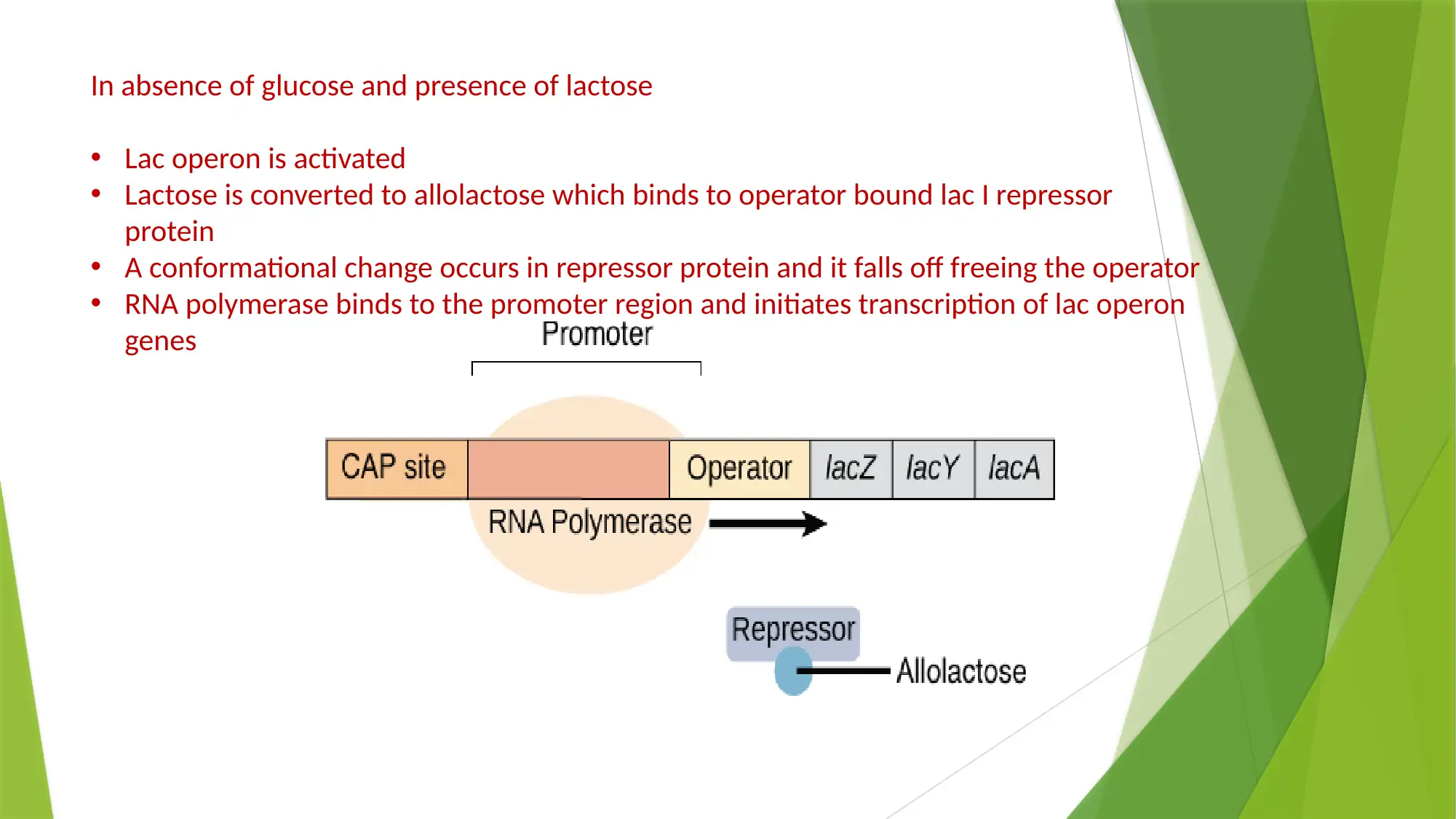
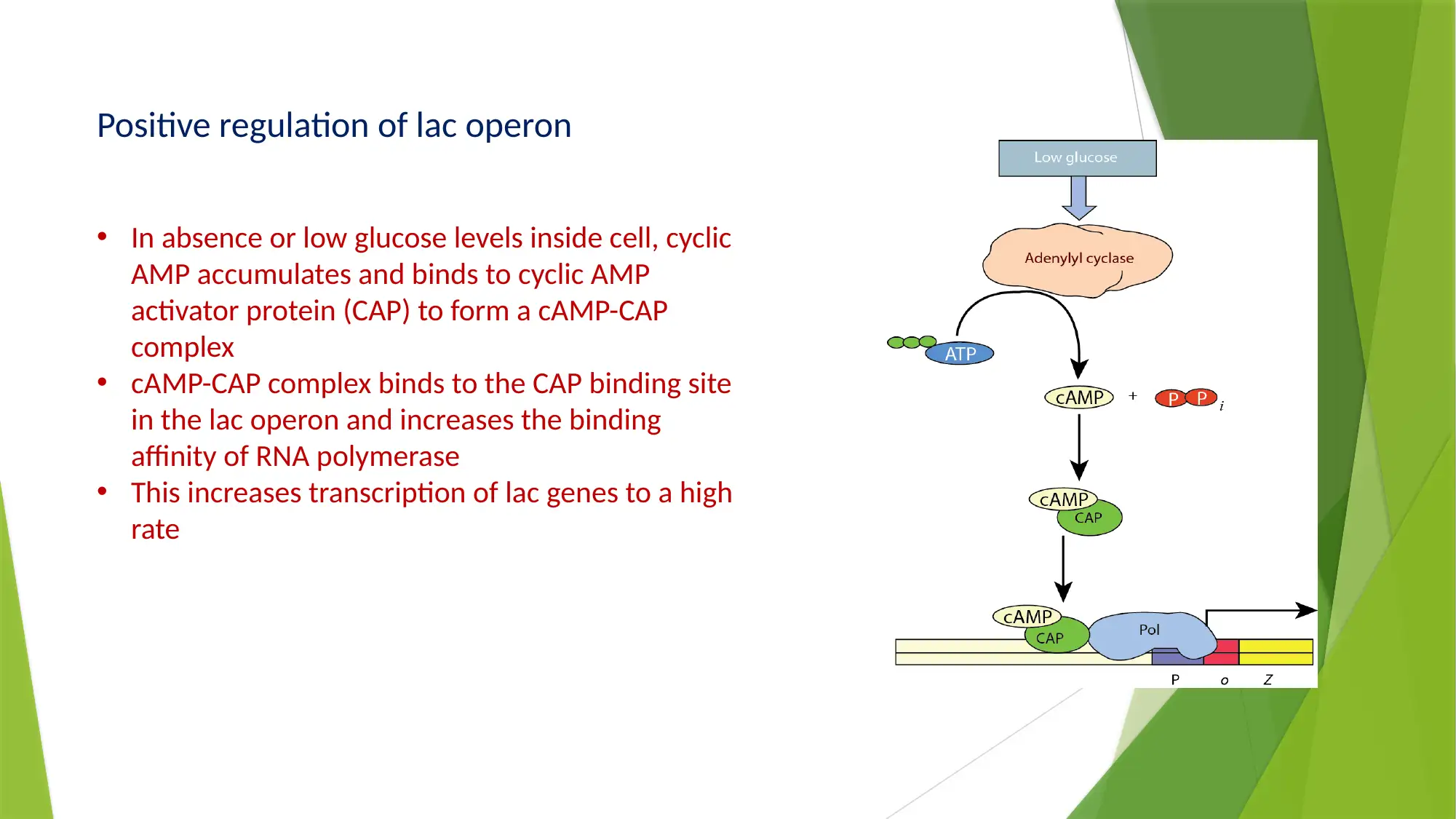
This essay provides a comprehensive comparison of gene expression and regulation in prokaryotic and eukaryotic systems. It begins by highlighting the similarities in gene structure, particularly the promoter regions, while emphasizing the compartmentalized nature of eukaryotic gene expression. The essay details the structure of prokaryotic genes, including open reading frames, promoter sequences, and regulatory elements like operators, enhancers, and silencers. It explains the function of the lac operon as an example of inducible gene regulation. The essay then transitions to eukaryotic gene structure, discussing promoter elements, chromatin remodeling, and pre-mRNA processing, including 5’ capping, 3’ polyadenylation, and alternative splicing. Key differences are highlighted, such as the monocistronic nature of eukaryotic transcripts and the role of chromatin structure. The essay concludes by summarizing the key differences in gene regulation and expression between the two systems, emphasizing the complexity of eukaryotic gene expression and the mechanisms that contribute to protein diversity.
1 out of 23








![[object Object]](/_next/static/media/star-bottom.7253800d.svg)