Glycine Max Sequence Alignment and Phylogenetic Tree Analysis Report
VerifiedAdded on 2022/08/19
|5
|469
|36
Report
AI Summary
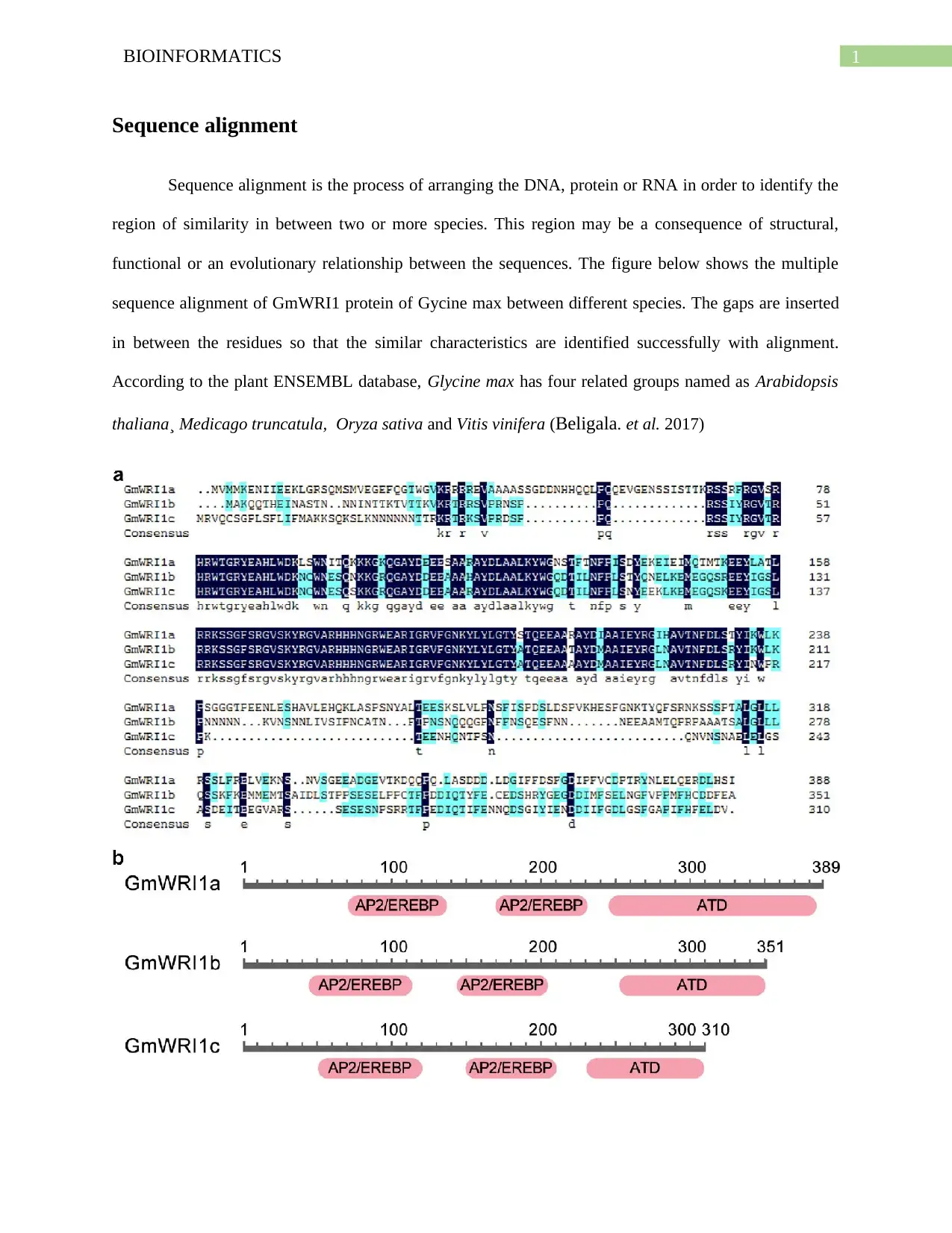
This report provides a bioinformatics analysis of Glycine max, focusing on sequence alignment and phylogenetic tree construction. The report examines the process of aligning DNA, protein, or RNA sequences to identify regions of similarity between species, using tools like MSA (Clustal W). The analysis includes a multiple sequence alignment of GmWRI1 protein and CLE protein of Glycine max, comparing it with sequences from related species found in the plant ENSEMBL database such as Arabidopsis thaliana, Medicago truncatula, Oryza sativa, and Vitis vinifera. The phylogenetic tree illustrates the evolutionary relationships between these species. Homology searches using BLAST tools and Hidden Markov Models are also discussed, revealing the mapping of SWEET genes to soybean chromosomes. Furthermore, the report highlights the expression profiles of SWEET genes across different developmental stages and tissues, indicating their association with reproductive tissue development. The analysis contributes to understanding the genetic makeup and evolutionary relationships of Glycine max.
1 out of 5








![[object Object]](/_next/static/media/star-bottom.7253800d.svg)