Genetic and Protein Analysis of H1N1 using BLAST and ExPASy
VerifiedAdded on 2022/09/27
|10
|3659
|17
Homework Assignment
AI Summary
This assignment focuses on the bioinformatics analysis of H1N1 influenza virus, utilizing BLAST and ExPASy tools to investigate genetic and protein variations, including mutations that confer resistance to antiviral medications. The assignment begins with an introduction to influenza and the H1N1 strain, followed by a background on the virus's nomenclature and the role of Hemagglutinin and Neuraminidase. The core of the assignment involves using BLAST to compare H1N1 sequences, align nucleotide and protein sequences, and identify mutations. The assignment is divided into parts, each involving specific BLAST searches and questions. Part A introduces BLASTn, comparing two non-resistant H1N1 strains to analyze nucleotide differences. Part B involves collecting information about the sequences, including collection dates, hosts, and protein IDs. Part C utilizes blastp to align protein sequences and identify amino acid differences and potential resistance-conferring variations. Part D extends the analysis by aligning multiple protein sequences, including both normal and mutant strains, to draw conclusions about the nature of mutations leading to antiviral resistance. The assignment aims to provide a comprehensive understanding of how bioinformatics tools are used to study viral evolution and drug resistance.

Using BLAST and ExPASy for Genetic and Protein analysis of H1N1 variability, including
mutations that confer resistance to antiviral medications.
Introduction: Influenza is an infection of the upper respiratory tract that causes sickness and
death and widespread outbreaks also cause a significant economic impact as well. The H1N1
influenza A pandemic of 2009 caused millions of people to become sick, hundreds of thousands
to be hospitalized and thousands of deaths in the United States alone1. The new flu strain
contained genes from influenza viruses from avian (bird), swine (pig) and human. New strains
often cause more infection because our immune systems have been conditioned to respond to
previously encountered strains. When strains mutate it allows them to more easily slip by our
body’s defenses. In addition, mutation can make previous vaccines and antiviral medications
ineffective against them.
Background: Most of us are familiar now with the nomenclature “H1N1” but few of us know
the meaning of these letters and numbers. Scientists label flu viruses based on the presence of
two antigens on their surface. The first letter, “H” refers to the type of Hemagglutinin present on
the virus’ surface. This molecule is primarily responsible for the virus’ ability to infect cells.
The second letter, “N”, refers to the type of Neuraminidase present. This molecule helps the
replicated virus leave from the cell in which is has replicated, allowing the many copies to infect
more cells. These two proteins together determine much about the virus, including, in general,
which species the virus can infect and the virulence of the particular strain. Even though there
are many different strains of flu virus, they infect mammals and birds and there remains a lot of
similarity between them. These similarities can be exploited to develop vaccines and/or antiviral
drug therapies that could be used against a wide array of similar viruses. Between 1999 and
2002 two new antiviral drugs were introduced into the population: zanamivir and oseltamivir.
These same drugs were used to treat patients with H1N1. In just about 10 years it is possible for
flu viruses to evolve resistance to these drugs. In order to monitor the potential development of
resistant strains the Neuraminidase Inhibitor Susceptibility Network was established.
Directions: You are a member of the Neuraminidase Inhibitor Susceptibility Network and the
World Health Organization has contacted you with information about a new viral strain that
might have resistance to the antiviral drugs zanamivir and oseltamivir (Tamiflu). In this case
study you will:
•Use BLAST to compare H1N1 sequences to observe conserved and variable regions
•Align nucleotide sequences of normal strains of H1N1
•Align multiple nucleotide and protein sequences to look for mutations.
•Compare the newly identified strain and hypothesize its resistance based on its mutations
Part A: Introduction to blastn. Listed below are the identifying numbers for several “normal”
(non-resistant) strains of H1N1 that were sequenced during the 2009 pandemic. In part A of this
activity you will use online database tools to look at virus variation that does not confer antiviral
resistance.
Step 1: Go to the National Center for Biotechnology Information’s website at:
http://www.ncbi.nlm.nih.gov. From the menu at the top right, click on BLAST. BLAST stands
for “Basic Local Alignment Search Tool” and it is one of the many web-based applications
mutations that confer resistance to antiviral medications.
Introduction: Influenza is an infection of the upper respiratory tract that causes sickness and
death and widespread outbreaks also cause a significant economic impact as well. The H1N1
influenza A pandemic of 2009 caused millions of people to become sick, hundreds of thousands
to be hospitalized and thousands of deaths in the United States alone1. The new flu strain
contained genes from influenza viruses from avian (bird), swine (pig) and human. New strains
often cause more infection because our immune systems have been conditioned to respond to
previously encountered strains. When strains mutate it allows them to more easily slip by our
body’s defenses. In addition, mutation can make previous vaccines and antiviral medications
ineffective against them.
Background: Most of us are familiar now with the nomenclature “H1N1” but few of us know
the meaning of these letters and numbers. Scientists label flu viruses based on the presence of
two antigens on their surface. The first letter, “H” refers to the type of Hemagglutinin present on
the virus’ surface. This molecule is primarily responsible for the virus’ ability to infect cells.
The second letter, “N”, refers to the type of Neuraminidase present. This molecule helps the
replicated virus leave from the cell in which is has replicated, allowing the many copies to infect
more cells. These two proteins together determine much about the virus, including, in general,
which species the virus can infect and the virulence of the particular strain. Even though there
are many different strains of flu virus, they infect mammals and birds and there remains a lot of
similarity between them. These similarities can be exploited to develop vaccines and/or antiviral
drug therapies that could be used against a wide array of similar viruses. Between 1999 and
2002 two new antiviral drugs were introduced into the population: zanamivir and oseltamivir.
These same drugs were used to treat patients with H1N1. In just about 10 years it is possible for
flu viruses to evolve resistance to these drugs. In order to monitor the potential development of
resistant strains the Neuraminidase Inhibitor Susceptibility Network was established.
Directions: You are a member of the Neuraminidase Inhibitor Susceptibility Network and the
World Health Organization has contacted you with information about a new viral strain that
might have resistance to the antiviral drugs zanamivir and oseltamivir (Tamiflu). In this case
study you will:
•Use BLAST to compare H1N1 sequences to observe conserved and variable regions
•Align nucleotide sequences of normal strains of H1N1
•Align multiple nucleotide and protein sequences to look for mutations.
•Compare the newly identified strain and hypothesize its resistance based on its mutations
Part A: Introduction to blastn. Listed below are the identifying numbers for several “normal”
(non-resistant) strains of H1N1 that were sequenced during the 2009 pandemic. In part A of this
activity you will use online database tools to look at virus variation that does not confer antiviral
resistance.
Step 1: Go to the National Center for Biotechnology Information’s website at:
http://www.ncbi.nlm.nih.gov. From the menu at the top right, click on BLAST. BLAST stands
for “Basic Local Alignment Search Tool” and it is one of the many web-based applications
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

available to investigate, analyze, research and identify genes and proteins of interest. The
applications of BLAST and other tools are almost limitless! All of these resources are free and
open to the public. This particular site is maintained by the National Institutes of Health (NIH).
Step 2: Choose nucleotide blast from the menu of choices under the heading “Basic BLAST”.
To become familiar with BLAST output we will compare the nucleotide sequences of two strains
of H1N1. Enter this accession number in the box, CY056295. Give your job the title, “align two
non-resistant H1N1 strains”.
Step 3: Click on the box in front of “Align two or more sequences”. This will open a new box.
Enter accession number CY062530 in this box.
Step 4: At the bottom section called “Program Selection” select the choice “Highly similar
sequences (megablast). Then click “show results in a new window” so we can refer back to our
search criteria if needed.
Step 5: Click the “BLAST” button. Your search will show up in a new window or tab and if
you scroll down you can see the aligned nucleotides for the two neuraminidase genes in our two
strains.
Answer the questions for Part A once the BLASTN search is finished.
Part A Questions:
1. What are the lengths of the two sequences used in this comparison?
Query: _____1410____ nucleotides Subject: __1421_______ nucleotides
2. Now that the sequences are aligned for best fit, how many differences can be found between
the two aligned regions? (count them!)
____11_____ nucleotides
3. Since every 3 bases codes for 1 amino acid, what is the maximum number of amino acids in
the protein that would be made from the shorter of the two sequences?
____470_____ amino acids
4. Will every nucleotide difference result in an amino acid difference? Explain your answer.
Yes, nucleotide difference leads to an amino acid difference because 3 nucleotides codes for one
amino acid. Hence, if there is a deletion of nucleotide would cause a difference between
nucleotide and amino acids.
5. Since neither of these strains is resistant to anti-viral therapies, what can be concluded about
the variation in the gene for neuraminidase between these two strains?
applications of BLAST and other tools are almost limitless! All of these resources are free and
open to the public. This particular site is maintained by the National Institutes of Health (NIH).
Step 2: Choose nucleotide blast from the menu of choices under the heading “Basic BLAST”.
To become familiar with BLAST output we will compare the nucleotide sequences of two strains
of H1N1. Enter this accession number in the box, CY056295. Give your job the title, “align two
non-resistant H1N1 strains”.
Step 3: Click on the box in front of “Align two or more sequences”. This will open a new box.
Enter accession number CY062530 in this box.
Step 4: At the bottom section called “Program Selection” select the choice “Highly similar
sequences (megablast). Then click “show results in a new window” so we can refer back to our
search criteria if needed.
Step 5: Click the “BLAST” button. Your search will show up in a new window or tab and if
you scroll down you can see the aligned nucleotides for the two neuraminidase genes in our two
strains.
Answer the questions for Part A once the BLASTN search is finished.
Part A Questions:
1. What are the lengths of the two sequences used in this comparison?
Query: _____1410____ nucleotides Subject: __1421_______ nucleotides
2. Now that the sequences are aligned for best fit, how many differences can be found between
the two aligned regions? (count them!)
____11_____ nucleotides
3. Since every 3 bases codes for 1 amino acid, what is the maximum number of amino acids in
the protein that would be made from the shorter of the two sequences?
____470_____ amino acids
4. Will every nucleotide difference result in an amino acid difference? Explain your answer.
Yes, nucleotide difference leads to an amino acid difference because 3 nucleotides codes for one
amino acid. Hence, if there is a deletion of nucleotide would cause a difference between
nucleotide and amino acids.
5. Since neither of these strains is resistant to anti-viral therapies, what can be concluded about
the variation in the gene for neuraminidase between these two strains?

It can be concluded that Neuraminidase (NA) is a surface proteins of influenza A virus, and a
variation in the gene is observed that plays an essential role in inoculation beside influenza
infection in addition to is predictable as imperative therapeutic objective. Hence, the genetic in
addition to antigenic variations besides exchanges can stimulates the effectiveness of vaccine in
addition to modification viral understanding to NA inhibitors (NAIs) (Göktepe & Kodaz, 2018).
Part B: Collecting Info on your sequences. There are a lot of details recorded about the
sequences that are uploaded to the GenBank database. In Part B of this activity we will collect
information about our strains, including a reference number to their translated amino acid
(protein) sequence. We will use these sequences in Pact C of this activity.
Step 1: From the top of the BLAST results page, click on the link after “Query ID”. This will
bring up all of the information about this sequenced gene. Look over the information and answer
the questions:
Part B Questions:
1. When was this sample collected?
The sample was collect on 7th March 2010.
2. Who was the host of this virus? Be as specific as possible.
The host virus is Influenza A virus (A/Henan/1/2010(H1N1))
3. From which country was this host?
The country China was its host.
4. If you scroll down to the bottom you can find the entire sequence and translated amino acid
sequence. See appendix A or http://www.bio.davidson.edu/courses/genomics/jmol/aatable.html
for a list of amino acid abbreviations. Write down the protein_id
number: ____ ADD52538.1" _______________
Step 2: Use the back button of the web browser to return to your BLAST results. Scroll down
and click on the accession number of our subject sequence, CY062530.1. Answer the same 4
questions about this viral strain.
1. When was this sample collected?
The sample was collected on 4th May 2010.
2. Who was the host of this virus? Be as specific as possible.
The host of this virus was Guadalajara, Jalisco, Mexico
variation in the gene is observed that plays an essential role in inoculation beside influenza
infection in addition to is predictable as imperative therapeutic objective. Hence, the genetic in
addition to antigenic variations besides exchanges can stimulates the effectiveness of vaccine in
addition to modification viral understanding to NA inhibitors (NAIs) (Göktepe & Kodaz, 2018).
Part B: Collecting Info on your sequences. There are a lot of details recorded about the
sequences that are uploaded to the GenBank database. In Part B of this activity we will collect
information about our strains, including a reference number to their translated amino acid
(protein) sequence. We will use these sequences in Pact C of this activity.
Step 1: From the top of the BLAST results page, click on the link after “Query ID”. This will
bring up all of the information about this sequenced gene. Look over the information and answer
the questions:
Part B Questions:
1. When was this sample collected?
The sample was collect on 7th March 2010.
2. Who was the host of this virus? Be as specific as possible.
The host virus is Influenza A virus (A/Henan/1/2010(H1N1))
3. From which country was this host?
The country China was its host.
4. If you scroll down to the bottom you can find the entire sequence and translated amino acid
sequence. See appendix A or http://www.bio.davidson.edu/courses/genomics/jmol/aatable.html
for a list of amino acid abbreviations. Write down the protein_id
number: ____ ADD52538.1" _______________
Step 2: Use the back button of the web browser to return to your BLAST results. Scroll down
and click on the accession number of our subject sequence, CY062530.1. Answer the same 4
questions about this viral strain.
1. When was this sample collected?
The sample was collected on 4th May 2010.
2. Who was the host of this virus? Be as specific as possible.
The host of this virus was Guadalajara, Jalisco, Mexico
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

3. From which country was this host?
Mexico country is the host of this virus
4. If you scroll down to the bottom you can find the entire sequence and translated amino acid
sequence. See appendix A or http://www.bio.davidson.edu/courses/genomics/jmol/aatable.html
for a list of amino acid abbreviations. Write down the protein_id
number: _____ ADG27998.1" ______________
Part C: Using blastp. In Part C of this activity we will use blastp to align two protein sequences.
Step 1: Navigate back to the BLAST input page. From the tabs at the top click on “blastp”. If
you have closed this window, go back to http://www.ncbi.nlm.nih.gov. From here choose “blast”
and then “protein blast”.
Step 2: Input the first of the two protein id numbers from Part B into the box. Click on “align
two or more sequences” and enter the second protein reference number in the second box.
Step 3: Choose “show results in a new window” and click the BLAST button. The blastp output
will show a large number of identifiers listed below the query sequence. The reason for the large
number of entries is because we are searching against the non-redundant protein database (nr).
As the database name suggests, NCBI tries to collapse entries in GenBank with the same
sequence into a single record and then simply append the redundant sequences in the description.
Step 4: Scroll down to the bottom to see the two sequences aligned. It is difficult to look for
amino acid differences in this view. To change the view to make it easier, scroll to the very top.
Click “Formatting options”. In the “alignment view” pull down menu, choose “query-anchored
with dots for identities”. Click the blue “reformat” button to see the changed view.
Step 5: Scroll back all the way to the bottom. Now you can see the identical amino acids
marked with a dot, and the differences are marked with their respective amino acid letters.
Answer the questions for Part C once the blastp search is complete.
Part C Questions:
1. How many amino acid differences are there between the two sequences?___two
2. Scientists use the single letter amino acid abbreviations and their position number to label and
refer to specific amino acids. Record the amino acid differences below:
Position 274: Query ___Y___ Position 453: Query __V____
Subject __H___ Subject _____M_
Mexico country is the host of this virus
4. If you scroll down to the bottom you can find the entire sequence and translated amino acid
sequence. See appendix A or http://www.bio.davidson.edu/courses/genomics/jmol/aatable.html
for a list of amino acid abbreviations. Write down the protein_id
number: _____ ADG27998.1" ______________
Part C: Using blastp. In Part C of this activity we will use blastp to align two protein sequences.
Step 1: Navigate back to the BLAST input page. From the tabs at the top click on “blastp”. If
you have closed this window, go back to http://www.ncbi.nlm.nih.gov. From here choose “blast”
and then “protein blast”.
Step 2: Input the first of the two protein id numbers from Part B into the box. Click on “align
two or more sequences” and enter the second protein reference number in the second box.
Step 3: Choose “show results in a new window” and click the BLAST button. The blastp output
will show a large number of identifiers listed below the query sequence. The reason for the large
number of entries is because we are searching against the non-redundant protein database (nr).
As the database name suggests, NCBI tries to collapse entries in GenBank with the same
sequence into a single record and then simply append the redundant sequences in the description.
Step 4: Scroll down to the bottom to see the two sequences aligned. It is difficult to look for
amino acid differences in this view. To change the view to make it easier, scroll to the very top.
Click “Formatting options”. In the “alignment view” pull down menu, choose “query-anchored
with dots for identities”. Click the blue “reformat” button to see the changed view.
Step 5: Scroll back all the way to the bottom. Now you can see the identical amino acids
marked with a dot, and the differences are marked with their respective amino acid letters.
Answer the questions for Part C once the blastp search is complete.
Part C Questions:
1. How many amino acid differences are there between the two sequences?___two
2. Scientists use the single letter amino acid abbreviations and their position number to label and
refer to specific amino acids. Record the amino acid differences below:
Position 274: Query ___Y___ Position 453: Query __V____
Subject __H___ Subject _____M_
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

3. Use Appendix A or http://www.bio.davidson.edu/courses/genomics/jmol/aatable.html to
record what change in the properties of amino acids has occurred at each variation site.
Site 274:_________Tyr____-->____His__________ Site 453:____Val_________--
>_________Met_____
4. Even though we know these strains do not cause anti-viral resistance, which amino acid
variation is more likely to confer resistance? Why?
Variation is histidine(His) would lead to resistance as it is having aromatic ring. Break in the
aromatic ring would create steric hindrance thereby causing resistance.
5. What can be concluded about this variation?
This variation might have cause due to some mutation or development of pathogenic activity.
Hence variation effect would lead serious effect if it infects the human.
Part D: Aligning multiple protein sequences. In Part D of this activity we will align multiple
neuraminidase protein sequences to find the nature of the mutation that confers resistance to the
two antiviral drugs, zanamivir and oseltamivir.
Step 1: Navigate back to the blastp tool (see part C). Keep the query sequence the same as in
Part C (ADD52538.1). Now click align two or more sequences and enter all of the following
numbers, each followed by the enter/return key. Be sure to keep them in order so we will know
which ones are resistant.
4 normal strains:
ADG27998
ADB98137
ACT67242
BAJ05804
and 4 mutant strains:
ADB98138
ADF97837
ACT10319
ADG28013
Step 2: Choose “Show results in a new window” and click the BLAST button.
Step 3: The default view for the results will show EACH sequence compared to the query
sequence. We want to see them all together. At the top, change the formatting options to “query
anchored with dots for identities”. Don’t forget to click the “Reformat” button for your changes
to take effect. Then scroll down to view all of your amino acid sequences aligned. Imagine
doing this analysis without a computer!
Part D Questions:
record what change in the properties of amino acids has occurred at each variation site.
Site 274:_________Tyr____-->____His__________ Site 453:____Val_________--
>_________Met_____
4. Even though we know these strains do not cause anti-viral resistance, which amino acid
variation is more likely to confer resistance? Why?
Variation is histidine(His) would lead to resistance as it is having aromatic ring. Break in the
aromatic ring would create steric hindrance thereby causing resistance.
5. What can be concluded about this variation?
This variation might have cause due to some mutation or development of pathogenic activity.
Hence variation effect would lead serious effect if it infects the human.
Part D: Aligning multiple protein sequences. In Part D of this activity we will align multiple
neuraminidase protein sequences to find the nature of the mutation that confers resistance to the
two antiviral drugs, zanamivir and oseltamivir.
Step 1: Navigate back to the blastp tool (see part C). Keep the query sequence the same as in
Part C (ADD52538.1). Now click align two or more sequences and enter all of the following
numbers, each followed by the enter/return key. Be sure to keep them in order so we will know
which ones are resistant.
4 normal strains:
ADG27998
ADB98137
ACT67242
BAJ05804
and 4 mutant strains:
ADB98138
ADF97837
ACT10319
ADG28013
Step 2: Choose “Show results in a new window” and click the BLAST button.
Step 3: The default view for the results will show EACH sequence compared to the query
sequence. We want to see them all together. At the top, change the formatting options to “query
anchored with dots for identities”. Don’t forget to click the “Reformat” button for your changes
to take effect. Then scroll down to view all of your amino acid sequences aligned. Imagine
doing this analysis without a computer!
Part D Questions:

1. What conclusions can you draw from the information? Be as specific and complete in your
explanation as possible.
Mostly the variation is observed between H and Y (His and Tyr) for amino acid ranging from
241 to 300 and one variation observed between the range 421 to 469 between Met and Val. This
proved that the sequence range from 241 to 300 are unstable and it can also be assumed that Try
is more stable amino acid than His. There are no variation observed in other sequence range
(Lamb et al., 2019).
Part E: Analyzing a new mutant strain. You have been asked to analyze a new resistant strain
of H1N1. You have been provided with the nucleotide sequence. In Part D of this activity you
will translate the nucleotide sequence into an amino acid sequence to draw conclusions about the
nature of its mutation.
Step 1: Navigate to http://expasy.org/tools/
There are many tools available to researchers at this site maintained by the SIB (Swiss Institute
of Bioinformatics.) Scroll to the bottom and choose translate. It can be found in section labeled
DNA-->protein.
Step 2: Copy and paste the sequence from appendix B into the box.
Step 3: Click on translate sequence. This will translate the nucleotide sequence in all of the
possible “frames”, 3 from the top strand and 3 from the bottom strand.
Step 4: Examine the translated sequences. Your protein will not have many stop codons in the
middle of the sequence and it should start with a “Met” for Methionine
Once you have located the most likely reading frame, click on this frame number.
Step 5: You will see a new window with your sequence. Copy this protein sequence starting
with the earliest M and ending with the last letter before the STOP codon.
Step 6: Return to the BLASTp search page from Part D. Paste this new protein sequence into
the second box. Be sure it is separate from the last entry by a enter/return.
Step 7: Choose results in a new window and click BLAST.
Step 8: Reformat the results like you did in Part D to view all of the sequences aligned together.
Don’t forget to click the reformat button.
Part E Questions:
1. What conclusions can you draw from the aligned sequences of the wild-type strain, original
mutant strain and the new mutant strain? Be as specific as possible in your analysis. Be sure to
include where researchers should focus their study in the future to further analyze the nature of
this new mutant strain.
Variation is observed between 61 to 120 between original and new mutant (E and G), 121-180 (L
and S; D and V), 241 to 300 (Y and H) among wild-type and original mutant, 361 to 420 (V and
K) among wild-type and new mutant and 421 to 469 (V and M) among wild-type or orginial
mutant. The conclusion can be drawn is that new mutant sequence is highly similar with the
original mutant strain ("UniProt: a worldwide hub of protein knowledge", 2018). Their similarity
explanation as possible.
Mostly the variation is observed between H and Y (His and Tyr) for amino acid ranging from
241 to 300 and one variation observed between the range 421 to 469 between Met and Val. This
proved that the sequence range from 241 to 300 are unstable and it can also be assumed that Try
is more stable amino acid than His. There are no variation observed in other sequence range
(Lamb et al., 2019).
Part E: Analyzing a new mutant strain. You have been asked to analyze a new resistant strain
of H1N1. You have been provided with the nucleotide sequence. In Part D of this activity you
will translate the nucleotide sequence into an amino acid sequence to draw conclusions about the
nature of its mutation.
Step 1: Navigate to http://expasy.org/tools/
There are many tools available to researchers at this site maintained by the SIB (Swiss Institute
of Bioinformatics.) Scroll to the bottom and choose translate. It can be found in section labeled
DNA-->protein.
Step 2: Copy and paste the sequence from appendix B into the box.
Step 3: Click on translate sequence. This will translate the nucleotide sequence in all of the
possible “frames”, 3 from the top strand and 3 from the bottom strand.
Step 4: Examine the translated sequences. Your protein will not have many stop codons in the
middle of the sequence and it should start with a “Met” for Methionine
Once you have located the most likely reading frame, click on this frame number.
Step 5: You will see a new window with your sequence. Copy this protein sequence starting
with the earliest M and ending with the last letter before the STOP codon.
Step 6: Return to the BLASTp search page from Part D. Paste this new protein sequence into
the second box. Be sure it is separate from the last entry by a enter/return.
Step 7: Choose results in a new window and click BLAST.
Step 8: Reformat the results like you did in Part D to view all of the sequences aligned together.
Don’t forget to click the reformat button.
Part E Questions:
1. What conclusions can you draw from the aligned sequences of the wild-type strain, original
mutant strain and the new mutant strain? Be as specific as possible in your analysis. Be sure to
include where researchers should focus their study in the future to further analyze the nature of
this new mutant strain.
Variation is observed between 61 to 120 between original and new mutant (E and G), 121-180 (L
and S; D and V), 241 to 300 (Y and H) among wild-type and original mutant, 361 to 420 (V and
K) among wild-type and new mutant and 421 to 469 (V and M) among wild-type or orginial
mutant. The conclusion can be drawn is that new mutant sequence is highly similar with the
original mutant strain ("UniProt: a worldwide hub of protein knowledge", 2018). Their similarity
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

index is very high however original sequence and ne mutant have variation. Hence it is provided
that there was mutation.
Part F: The anti-viral drugs zanamivir and oseltamivir target the active site of
the catalytic component of neuraminidase. This active site has several key amino acids that
function in the chemical reaction. In Part F of this activity you will research the new mutations
amino acid substitutions to draw conclusions about which mutation is the likely cause of the
antiviral resistance.
Step 1: Use Google to search for “PMCID: PMC1563878”. Click on the first result. This will
bring up the Journal of Virology article, “Importance of Neuraminidase Active-Site Residues to
the Neuraminidase Inhibitor Resistance of Influenza Viruses”.
Step 2: Scroll down about halfway looking for figure 1. Click to enlarge this figure. This
image shows the active site of neuraminidase and its interactions with its substrate, sialic acid
(yellow). Antiviral drugs, such as Tamiflu act by mimicking the structure of sialic acid and
blocking this active site.
Part F Questions:
1. List below the neuraminidase amino acids that seem to play important roles in binding of
sialic acid:
____Agr371_________ __Agr292___________ _____Glu277________
_______Arg152______
______Glu276_____Glu277__ ___Glu119__________ __Glu425___________
Arg118_____________
_____________ _____________ _____________ _____________
_____________ _____________ _____________ _____________
2. Which, if any, of these amino acids are mutated in our new strain?
If any of these amino acids are mutated, then the binding of sialic acid will not occur that would
further cause improper antibody linked clearance for pathogens, therefore the pathogenic activity
will increase (Chong et al., 2018)
3. How should researchers use this new information?
Researcher can use this data for site-directed mutagenesis in addition to reverse genetics which are
plasmid base thereby permitting only the analysis of the impact of preserved residues on NAI resistance
however researcher mist also keeps in mind that viability assay for the recombinant viruses that are
carrying mutations for the residues need to analyzed properly (Chong et al., 2018). This NAIs were
designed depending on the structure, where every modification can differentiate from the sialic acid and
natural substrate. Zanamivir is altered at C4, in the rea where sialic acid hydroxyl group is replaced with a
guanidine group.
that there was mutation.
Part F: The anti-viral drugs zanamivir and oseltamivir target the active site of
the catalytic component of neuraminidase. This active site has several key amino acids that
function in the chemical reaction. In Part F of this activity you will research the new mutations
amino acid substitutions to draw conclusions about which mutation is the likely cause of the
antiviral resistance.
Step 1: Use Google to search for “PMCID: PMC1563878”. Click on the first result. This will
bring up the Journal of Virology article, “Importance of Neuraminidase Active-Site Residues to
the Neuraminidase Inhibitor Resistance of Influenza Viruses”.
Step 2: Scroll down about halfway looking for figure 1. Click to enlarge this figure. This
image shows the active site of neuraminidase and its interactions with its substrate, sialic acid
(yellow). Antiviral drugs, such as Tamiflu act by mimicking the structure of sialic acid and
blocking this active site.
Part F Questions:
1. List below the neuraminidase amino acids that seem to play important roles in binding of
sialic acid:
____Agr371_________ __Agr292___________ _____Glu277________
_______Arg152______
______Glu276_____Glu277__ ___Glu119__________ __Glu425___________
Arg118_____________
_____________ _____________ _____________ _____________
_____________ _____________ _____________ _____________
2. Which, if any, of these amino acids are mutated in our new strain?
If any of these amino acids are mutated, then the binding of sialic acid will not occur that would
further cause improper antibody linked clearance for pathogens, therefore the pathogenic activity
will increase (Chong et al., 2018)
3. How should researchers use this new information?
Researcher can use this data for site-directed mutagenesis in addition to reverse genetics which are
plasmid base thereby permitting only the analysis of the impact of preserved residues on NAI resistance
however researcher mist also keeps in mind that viability assay for the recombinant viruses that are
carrying mutations for the residues need to analyzed properly (Chong et al., 2018). This NAIs were
designed depending on the structure, where every modification can differentiate from the sialic acid and
natural substrate. Zanamivir is altered at C4, in the rea where sialic acid hydroxyl group is replaced with a
guanidine group.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
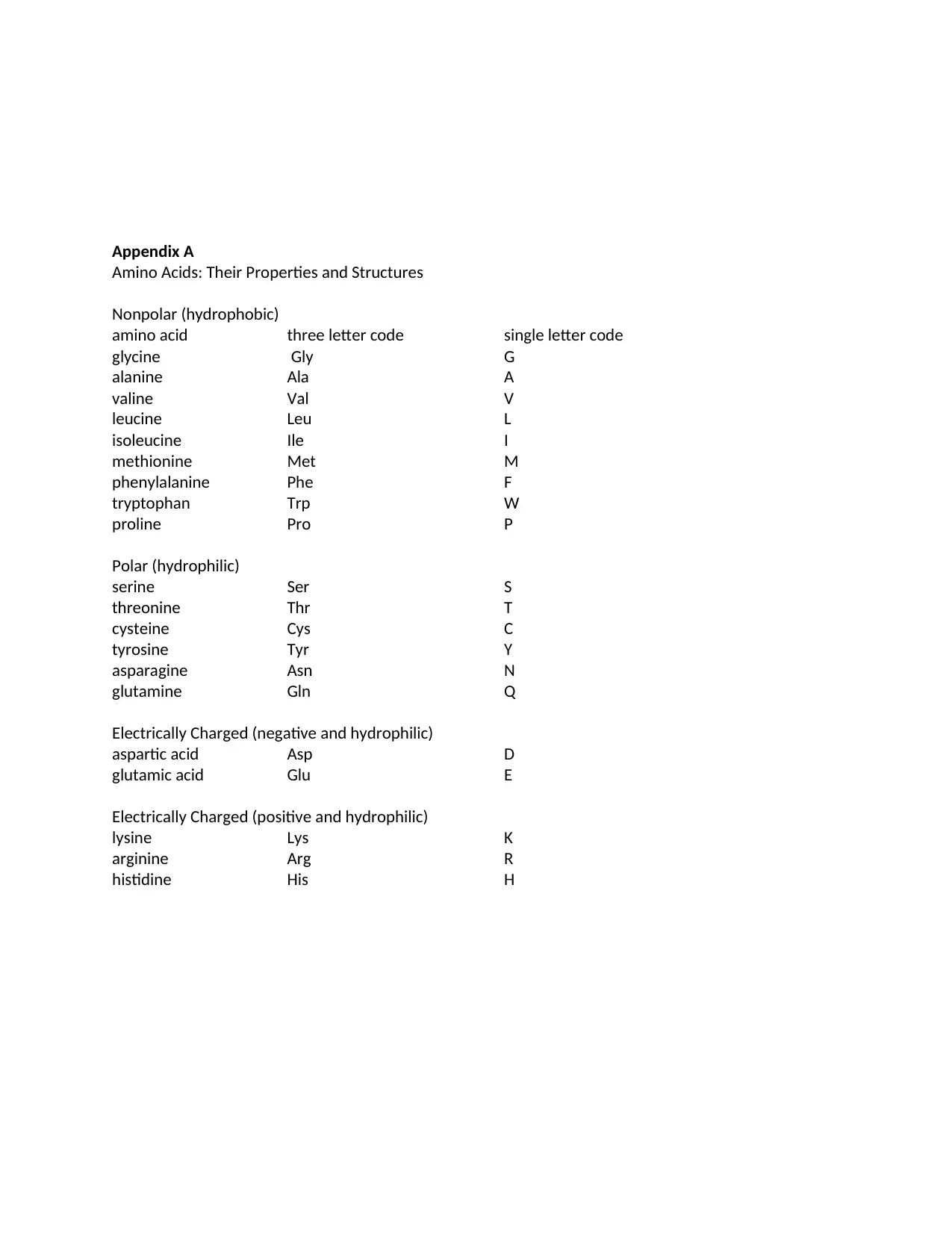
Appendix A
Amino Acids: Their Properties and Structures
Nonpolar (hydrophobic)
amino acid three letter code single letter code
glycine Gly G
alanine Ala A
valine Val V
leucine Leu L
isoleucine Ile I
methionine Met M
phenylalanine Phe F
tryptophan Trp W
proline Pro P
Polar (hydrophilic)
serine Ser S
threonine Thr T
cysteine Cys C
tyrosine Tyr Y
asparagine Asn N
glutamine Gln Q
Electrically Charged (negative and hydrophilic)
aspartic acid Asp D
glutamic acid Glu E
Electrically Charged (positive and hydrophilic)
lysine Lys K
arginine Arg R
histidine His H
Amino Acids: Their Properties and Structures
Nonpolar (hydrophobic)
amino acid three letter code single letter code
glycine Gly G
alanine Ala A
valine Val V
leucine Leu L
isoleucine Ile I
methionine Met M
phenylalanine Phe F
tryptophan Trp W
proline Pro P
Polar (hydrophilic)
serine Ser S
threonine Thr T
cysteine Cys C
tyrosine Tyr Y
asparagine Asn N
glutamine Gln Q
Electrically Charged (negative and hydrophilic)
aspartic acid Asp D
glutamic acid Glu E
Electrically Charged (positive and hydrophilic)
lysine Lys K
arginine Arg R
histidine His H

Appendix B
>gi|NewResistantH1N1|2010| agtttaaaat gaatccaaac caaaagataa taaccattgg ttcggtctgt atgacaattg
gaatggctaa cttaatatta caaattggaa acataatctc aatatggatt agccactcaa ttcaacttgg gaatcaaaat cagattgaaa
catgcaatca aagcgtcatt acttatgaaa acaacacttg ggtaaatcag acatatgtta acatcagcaa caccaacttt gctgctggac
agtcagtggt ttccgtgaaa ttagcgggca attcctctct ctgccctgtt agtggatggg ctatatacag taaagacaac agtataagaa
tcggttccaa gggggatgtg tttgtcataa ggggaccatt catatcatgt tcccccttgg aatgcagaac cttcttctcg actcaagggg
ccttgctaaa tgacaaacat tccaatggaa ccattaaagt taggagccca tatcgaaccc taatgagctg tcctattggt gaagttccct
ctccatacaa ctcaagattt gagtcagtcg cttggtcagc aagtgcttgt catgatggca tcaattggct aacaattgga atttctggcc
cagacaatgg ggcagtggct gtgttaaagt acaacggcat aataacagac actatcaaga gttggagaaa caatatattg agaacacaag
agtctgaatg tgcatgtgta aatggttctt gctttactgt aatgaccgat ggaccaagtg atggacaggc ctcatacaag atcttcagaa
tagaaaaggg aaagatagtc aaatcagtcg aaatgaatgc ccctaattat cactatgagg aatgctcctg ttatcctgat tctagtgaaa
tcacatgtgt gtgcagggat aactggcatg gctcgaatcg accgtgggtg tctttcaatc agaatctgga atatcagata ggatacatat
gcagtgggat tttcggagac aatccacgcc ctaatgataa gacaggcagt tgtggtccag tatcgtctaa tggagcaaat ggagtaaaag
gattttcatt caaatacggc aatggtgttt ggatagggag aactaaaagc attagttcaa gaaacggttt tgagatgatt tgggatccga
acggatggac tgggacagac aataacttct caataaagca agatatcgta ggaataaatg agtggtcagg atatagcggg agttttaagc
agcatccaga actaacaggg ctggattgta taagaccttg cttctgggtt gaactaatca gagggcgacc caaagagaac acaatctgga
ctagcgggag cagcatatcc ttttgcggtg taaacagtga cactgtgggt tggtcttggc cagacggtgc tgagttgcca tttaccattg
acaagtaatt tgttcaaaaa act
>gi|NewResistantH1N1|2010| agtttaaaat gaatccaaac caaaagataa taaccattgg ttcggtctgt atgacaattg
gaatggctaa cttaatatta caaattggaa acataatctc aatatggatt agccactcaa ttcaacttgg gaatcaaaat cagattgaaa
catgcaatca aagcgtcatt acttatgaaa acaacacttg ggtaaatcag acatatgtta acatcagcaa caccaacttt gctgctggac
agtcagtggt ttccgtgaaa ttagcgggca attcctctct ctgccctgtt agtggatggg ctatatacag taaagacaac agtataagaa
tcggttccaa gggggatgtg tttgtcataa ggggaccatt catatcatgt tcccccttgg aatgcagaac cttcttctcg actcaagggg
ccttgctaaa tgacaaacat tccaatggaa ccattaaagt taggagccca tatcgaaccc taatgagctg tcctattggt gaagttccct
ctccatacaa ctcaagattt gagtcagtcg cttggtcagc aagtgcttgt catgatggca tcaattggct aacaattgga atttctggcc
cagacaatgg ggcagtggct gtgttaaagt acaacggcat aataacagac actatcaaga gttggagaaa caatatattg agaacacaag
agtctgaatg tgcatgtgta aatggttctt gctttactgt aatgaccgat ggaccaagtg atggacaggc ctcatacaag atcttcagaa
tagaaaaggg aaagatagtc aaatcagtcg aaatgaatgc ccctaattat cactatgagg aatgctcctg ttatcctgat tctagtgaaa
tcacatgtgt gtgcagggat aactggcatg gctcgaatcg accgtgggtg tctttcaatc agaatctgga atatcagata ggatacatat
gcagtgggat tttcggagac aatccacgcc ctaatgataa gacaggcagt tgtggtccag tatcgtctaa tggagcaaat ggagtaaaag
gattttcatt caaatacggc aatggtgttt ggatagggag aactaaaagc attagttcaa gaaacggttt tgagatgatt tgggatccga
acggatggac tgggacagac aataacttct caataaagca agatatcgta ggaataaatg agtggtcagg atatagcggg agttttaagc
agcatccaga actaacaggg ctggattgta taagaccttg cttctgggtt gaactaatca gagggcgacc caaagagaac acaatctgga
ctagcgggag cagcatatcc ttttgcggtg taaacagtga cactgtgggt tggtcttggc cagacggtgc tgagttgcca tttaccattg
acaagtaatt tgttcaaaaa act
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

References
Chong, Y., Matsumoto, S., Kang, D., & Ikematsu, H. (2018). Consecutive influenza surveillance
of neuraminidase mutations and neuraminidase inhibitor resistance in Japan. Influenza
And Other Respiratory Viruses. https://doi.org/10.1111/irv.12624
Göktepe, Y., & Kodaz, H. (2018). Prediction of Protein-Protein Interactions Using An Effective
Sequence Based Combined Method. Neurocomputing, 303, 68-74.
https://doi.org/10.1016/j.neucom.2018.03.062
Lamb, J., Jarmolinska, A., Michel, M., Menéndez-Hurtado, D., Sulkowska, J., & Elofsson, A.
(2019). PconsFam: An Interactive Database of Structure Predictions of Pfam
Families. Journal Of Molecular Biology, 431(13), 2442-2448.
https://doi.org/10.1016/j.jmb.2019.01.047
UniProt: a worldwide hub of protein knowledge. (2018), 47(D1), D506-D515.
https://doi.org/10.1093/nar/gky1049
Chong, Y., Matsumoto, S., Kang, D., & Ikematsu, H. (2018). Consecutive influenza surveillance
of neuraminidase mutations and neuraminidase inhibitor resistance in Japan. Influenza
And Other Respiratory Viruses. https://doi.org/10.1111/irv.12624
Göktepe, Y., & Kodaz, H. (2018). Prediction of Protein-Protein Interactions Using An Effective
Sequence Based Combined Method. Neurocomputing, 303, 68-74.
https://doi.org/10.1016/j.neucom.2018.03.062
Lamb, J., Jarmolinska, A., Michel, M., Menéndez-Hurtado, D., Sulkowska, J., & Elofsson, A.
(2019). PconsFam: An Interactive Database of Structure Predictions of Pfam
Families. Journal Of Molecular Biology, 431(13), 2442-2448.
https://doi.org/10.1016/j.jmb.2019.01.047
UniProt: a worldwide hub of protein knowledge. (2018), 47(D1), D506-D515.
https://doi.org/10.1093/nar/gky1049
1 out of 10
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.



