Detailed Report on Human Carnitine Acetyltransferase: Structure & Role
VerifiedAdded on 2023/04/08
|6
|719
|79
Report
AI Summary
This report provides a detailed analysis of Human Carnitine Acetyltransferase, an enzyme crucial for metabolic maintenance. It identifies the protein, explains its role in catalyzing acyl group exchanges between carnitine and CoA, and describes its structural features, including its N and C domains, active site, and alpha-helices/beta-sheets composition. The report also contrasts Carnitine Acetyltransferase with haemoglobin, highlighting differences in domain numbers and amino acid composition, and discusses the protein's potential resistance to heat denaturation based on its structural properties. The document is available on Desklib, a platform offering a wealth of study resources for students.

PROTEIN ID 1NM8
0
0
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Table of Contents
Q1 - What is the name of the protein?.......................................................................................2
Q2- What does the protein do?..................................................................................................2
Q3 - What are the structural features of the protein?.................................................................2
Q4 - What are two features of your protein’s structure that make it different OR similar to
haemoglobin?.............................................................................................................................3
Q5 - Based on the structural properties of your protein, how resistant (or sensitive) would
your protein be to heat denaturation and why?..........................................................................4
References..................................................................................................................................5
1
Q1 - What is the name of the protein?.......................................................................................2
Q2- What does the protein do?..................................................................................................2
Q3 - What are the structural features of the protein?.................................................................2
Q4 - What are two features of your protein’s structure that make it different OR similar to
haemoglobin?.............................................................................................................................3
Q5 - Based on the structural properties of your protein, how resistant (or sensitive) would
your protein be to heat denaturation and why?..........................................................................4
References..................................................................................................................................5
1
Q1 - What is the name of the protein?
Human Carnitine Acetyltransferase
Q2- What does the protein do?
Carnitine Acetyltransferase performs a pivotal role in the maintenance of the metabolism of
the body. It is a family of enzymes that all catalyse similar reaction in which the acyl groups
are exchanges between carnitine and CoA or coenzyme A. Human Carnitine
Acetyltransferase, also identified as the Carnitine O-acetyltransferase is an enzyme coded by
the CRAT gene (Ncbi, 2019). Carnitine Acetyltransferase also acts as a fat metaboliser and is
found is both in the peroxisome and mitochondria. It plays the roles of a catalyst for the
reaction acetyl-CoA carnitine ⇌ CoA acetylcarnitine.
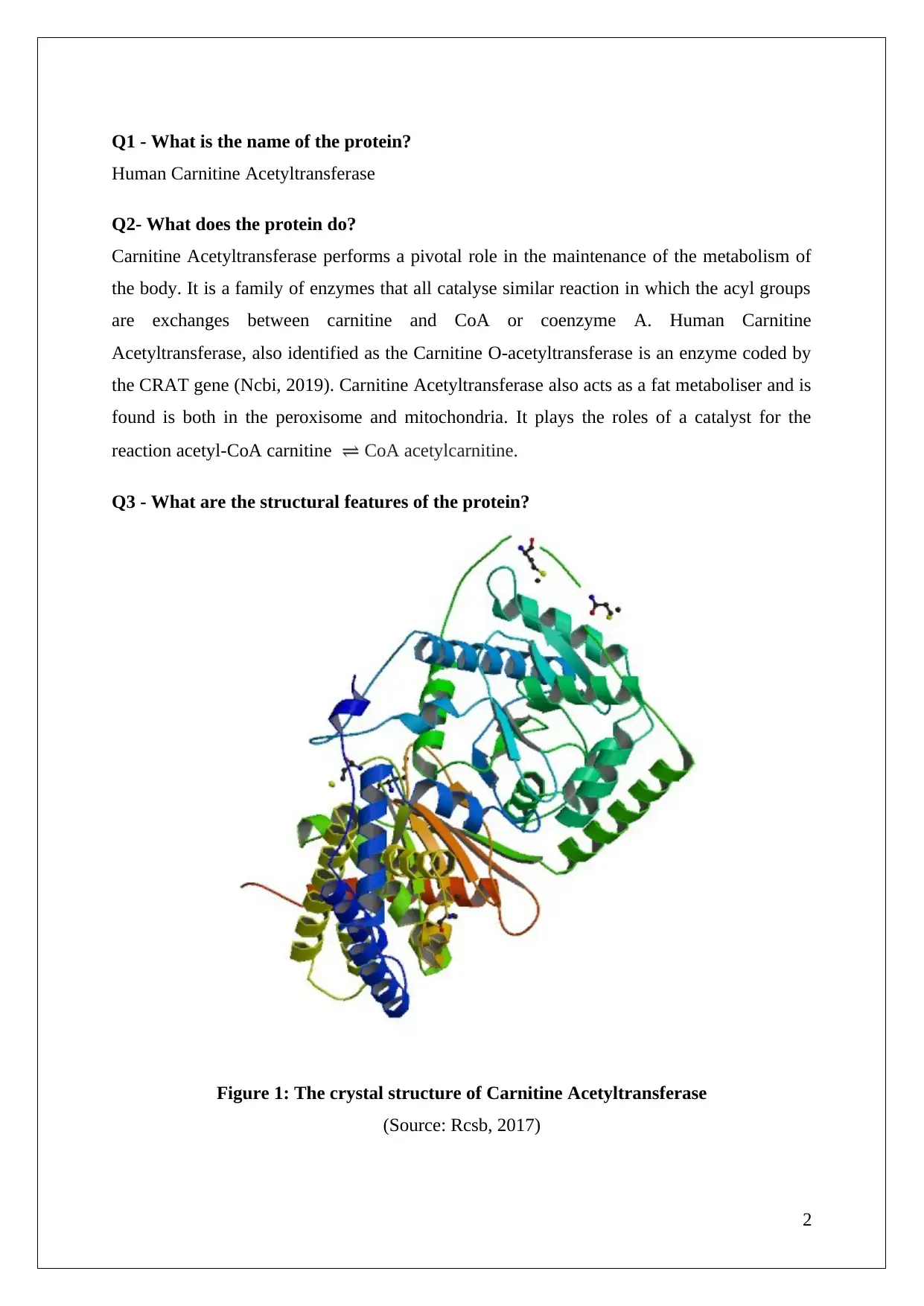
Q3 - What are the structural features of the protein?
Figure 1: The crystal structure of Carnitine Acetyltransferase
(Source: Rcsb, 2017)
2
Human Carnitine Acetyltransferase
Q2- What does the protein do?
Carnitine Acetyltransferase performs a pivotal role in the maintenance of the metabolism of
the body. It is a family of enzymes that all catalyse similar reaction in which the acyl groups
are exchanges between carnitine and CoA or coenzyme A. Human Carnitine
Acetyltransferase, also identified as the Carnitine O-acetyltransferase is an enzyme coded by
the CRAT gene (Ncbi, 2019). Carnitine Acetyltransferase also acts as a fat metaboliser and is
found is both in the peroxisome and mitochondria. It plays the roles of a catalyst for the
reaction acetyl-CoA carnitine ⇌ CoA acetylcarnitine.
Q3 - What are the structural features of the protein?
Figure 1: The crystal structure of Carnitine Acetyltransferase
(Source: Rcsb, 2017)
2
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

The molecular weight of carnitine acetyltransferase is 70kDa and is present in mitochondria
and the peroxisome. It comprises roughly 600 residues. Within the enzyme, two domains can
be identified, an N domain and a C domain. The N domain has eight stranded beta sheets
with eight alpha helices placed on both sides. The C domain has eleven alpha helices and six
stranded beta sheets. The two domains are arranged in a manner that a central narrow active
site shaped like a tunnel is originated. This appears to be a universal feature of the carnitine
acyltransferase family. The active site is His343 and it is located at the heart between the C
and N domains (Rcsb, 2017). The substrate can reach the site through two 15-18 A channels.
Through these channels, the substrate reaches the site at two opposite ends of the enzyme.
Substrates utilise one channel to reach the carnitine end while another for the CoA end.
Residues that form the carnitine binding site are found in both N and C domains. Carnitine is
required for catalysis and not really for the process of actual binding. The tertiary structure is
made up of 17 helices and 14 strands. They are further arranged into two alpha and beta
domains.
Q4 - What are two features of your protein’s structure that make it different OR
similar to haemoglobin?
Figure 2: Crystal structure of Haemoglobin
(Source: Rcsb, 2017)
Structural differences lie in the domain numbers. Haemoglobin molecule has 4 domains
whereas carnitine acetyltransferase has two. Carnitine acetyltransferase comprises of 20 alpha
helices and 16 beta strands while haemoglobin has dominant helical secondary structure
(Altamimi et al. 2018). Haemoglobin alpha chain is composed of 141 amino acids and the
3
and the peroxisome. It comprises roughly 600 residues. Within the enzyme, two domains can
be identified, an N domain and a C domain. The N domain has eight stranded beta sheets
with eight alpha helices placed on both sides. The C domain has eleven alpha helices and six
stranded beta sheets. The two domains are arranged in a manner that a central narrow active
site shaped like a tunnel is originated. This appears to be a universal feature of the carnitine
acyltransferase family. The active site is His343 and it is located at the heart between the C
and N domains (Rcsb, 2017). The substrate can reach the site through two 15-18 A channels.
Through these channels, the substrate reaches the site at two opposite ends of the enzyme.
Substrates utilise one channel to reach the carnitine end while another for the CoA end.
Residues that form the carnitine binding site are found in both N and C domains. Carnitine is
required for catalysis and not really for the process of actual binding. The tertiary structure is
made up of 17 helices and 14 strands. They are further arranged into two alpha and beta
domains.
Q4 - What are two features of your protein’s structure that make it different OR
similar to haemoglobin?
Figure 2: Crystal structure of Haemoglobin
(Source: Rcsb, 2017)
Structural differences lie in the domain numbers. Haemoglobin molecule has 4 domains
whereas carnitine acetyltransferase has two. Carnitine acetyltransferase comprises of 20 alpha
helices and 16 beta strands while haemoglobin has dominant helical secondary structure
(Altamimi et al. 2018). Haemoglobin alpha chain is composed of 141 amino acids and the
3
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

beta chain has 146 amino acids. In case of Carnitine acetyltransferase, the alpha chain has
616 amino acid.
Q5 - Based on the structural properties of your protein, how resistant (or sensitive)
would your protein be to heat denaturation and why?
The enzyme has a greater number of alpha helices than the number of beta strands. As a
result, it is likely to be granted better heat resisting qualities (van der Hoek et al.2018).
Therefore, like other enzymes in the human body, it can sustain temperatures up to 50 to 60
degree Celsius.
4
616 amino acid.
Q5 - Based on the structural properties of your protein, how resistant (or sensitive)
would your protein be to heat denaturation and why?
The enzyme has a greater number of alpha helices than the number of beta strands. As a
result, it is likely to be granted better heat resisting qualities (van der Hoek et al.2018).
Therefore, like other enzymes in the human body, it can sustain temperatures up to 50 to 60
degree Celsius.
4

References
Altamimi, T.R., Thomas, P.D., Darwesh, A.M., Fillmore, N., Mahmoud, M.U., Zhang, L.,
Gupta, A., Al Batran, R., Seubert, J.M. and Lopaschuk, G.D., (2018). Cytosolic carnitine
acetyltransferase as a source of cytosolic acetyl-CoA: a possible mechanism for regulation of
cardiac energy metabolism. Biochemical Journal, 475(5), pp.959-976.
Ncbi (2019), Carnitine O-acetyltransferase [Online], Available at:
https://www.ncbi.nlm.nih.gov/gene/1384 [Accessed 3 May 2019]
Rcsb (2017), Crystal structure of Carnitine Acetyltransferase [Online], Available at:
https://www.rcsb.org/structure/1NDB [Accessed 3 May 2019]
Rcsb, (2017) The Crystal Structure of Human Deoxyhaemoglobin [Online] Available
at:https://www.rcsb.org/structure/4hhb#entry-history [Accessed 3 May 2019]
van der Hoek, M.D., Madsen, O., Keijer, J. and van der Leij, F.R., (2018). Evolutionary
analysis of the carnitine-and choline acyltransferase suggests distinct evolution of CPT2
versus CPT1 and related variants. Biochimica et Biophysica Acta (BBA)-Molecular and Cell
Biology of Lipids, 1863(8), pp.909-918.
5
Altamimi, T.R., Thomas, P.D., Darwesh, A.M., Fillmore, N., Mahmoud, M.U., Zhang, L.,
Gupta, A., Al Batran, R., Seubert, J.M. and Lopaschuk, G.D., (2018). Cytosolic carnitine
acetyltransferase as a source of cytosolic acetyl-CoA: a possible mechanism for regulation of
cardiac energy metabolism. Biochemical Journal, 475(5), pp.959-976.
Ncbi (2019), Carnitine O-acetyltransferase [Online], Available at:
https://www.ncbi.nlm.nih.gov/gene/1384 [Accessed 3 May 2019]
Rcsb (2017), Crystal structure of Carnitine Acetyltransferase [Online], Available at:
https://www.rcsb.org/structure/1NDB [Accessed 3 May 2019]
Rcsb, (2017) The Crystal Structure of Human Deoxyhaemoglobin [Online] Available
at:https://www.rcsb.org/structure/4hhb#entry-history [Accessed 3 May 2019]
van der Hoek, M.D., Madsen, O., Keijer, J. and van der Leij, F.R., (2018). Evolutionary
analysis of the carnitine-and choline acyltransferase suggests distinct evolution of CPT2
versus CPT1 and related variants. Biochimica et Biophysica Acta (BBA)-Molecular and Cell
Biology of Lipids, 1863(8), pp.909-918.
5
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 6
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.


