Biology Assignment: Metabolic Processes in Cheese Production
VerifiedAdded on 2023/06/03
|6
|882
|416
Report
AI Summary
This biology assignment explores the metabolic processes involved in cheese production, focusing on the role of lactic acid bacteria (LAB) in fermentation. The solution details the biochemical reactions, including glycolysis, that occur during cheese ripening, acidification, and flavor development. The assignment explains how LAB, such as Lactococcus lactis, convert carbohydrates into lactic acid through anaerobic fermentation, affecting the final product's taste and preservation. It further elucidates the degradation of proteins, the role of enzymes like protease and rennet, and the formation of volatile compounds that contribute to the distinct flavors of different cheese varieties. The document also includes flowcharts and diagrams illustrating the metabolic pathways involved in cheese production and provides references to relevant research.

Running head: BIOLOGY ASSIGNMENT
BIOLOGY ASSIGNMENT
Name of the Student:
Name of the University:
Author Note:
BIOLOGY ASSIGNMENT
Name of the Student:
Name of the University:
Author Note:
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

1BIOLOGY ASSIGNMENT
Lactic acid bacteria which is popularly known as LAB is used in the fermentation
process. LAB help in the process of ripening and adds a characteristic flavour to the
fermented milk product. The fermentation process involves a series of metabolic effects that
includes acidification, breakdown of protein molecules and formation of volatile compounds.
The lactic acid bacteria leads to the formation of lactic acid through the metabolism of
carbohydrates to produce pyruvic acid. The procedure involved is known as anaerobic
fermentation. The biochemical process involves glycolysis to breakdown the carbohydrate
substrate into pyruvate which is further converted to lactic acid under the action of the
enzyme lactate dehydrogenase (Linares et al., 2013). The commonly used lactic acid bacteria
for the fermentation process includes, Lactococcus lactis which incorporates two subspecies,
namely lactis and cremoris (Linares et al., 2013).
The procedure begins with the degradation of the protein constituent in the starting
culture. The enzyme protease that is present in the initial culture degrades the protein
component. Rennet present in curd and plasmin present in milk help in the degradation of
protein and lead to the formation of amino acids and peptides. These components impart the
distinct flavour and taste properties to cheese. The free amino acids are converted to volatile
components by the action of the enzyme amino-acid converting enzyme (Linares et al.,
2013). On account of the difference in the strains of the LAB the regulating mechanism of the
amino acid converting enzyme is responsible for imparting different flavours to different
types of cheese. Further, it should be noted that, the metabolism of the lactic acid bacteria
helps in curdling and also effectively lowers the pH of the final product so as to prevent it
from the manifestation of other microbes.
The biochemical process of cheese production is elucidated as follows:
Lactic acid bacteria which is popularly known as LAB is used in the fermentation
process. LAB help in the process of ripening and adds a characteristic flavour to the
fermented milk product. The fermentation process involves a series of metabolic effects that
includes acidification, breakdown of protein molecules and formation of volatile compounds.
The lactic acid bacteria leads to the formation of lactic acid through the metabolism of
carbohydrates to produce pyruvic acid. The procedure involved is known as anaerobic
fermentation. The biochemical process involves glycolysis to breakdown the carbohydrate
substrate into pyruvate which is further converted to lactic acid under the action of the
enzyme lactate dehydrogenase (Linares et al., 2013). The commonly used lactic acid bacteria
for the fermentation process includes, Lactococcus lactis which incorporates two subspecies,
namely lactis and cremoris (Linares et al., 2013).
The procedure begins with the degradation of the protein constituent in the starting
culture. The enzyme protease that is present in the initial culture degrades the protein
component. Rennet present in curd and plasmin present in milk help in the degradation of
protein and lead to the formation of amino acids and peptides. These components impart the
distinct flavour and taste properties to cheese. The free amino acids are converted to volatile
components by the action of the enzyme amino-acid converting enzyme (Linares et al.,
2013). On account of the difference in the strains of the LAB the regulating mechanism of the
amino acid converting enzyme is responsible for imparting different flavours to different
types of cheese. Further, it should be noted that, the metabolism of the lactic acid bacteria
helps in curdling and also effectively lowers the pH of the final product so as to prevent it
from the manifestation of other microbes.
The biochemical process of cheese production is elucidated as follows:
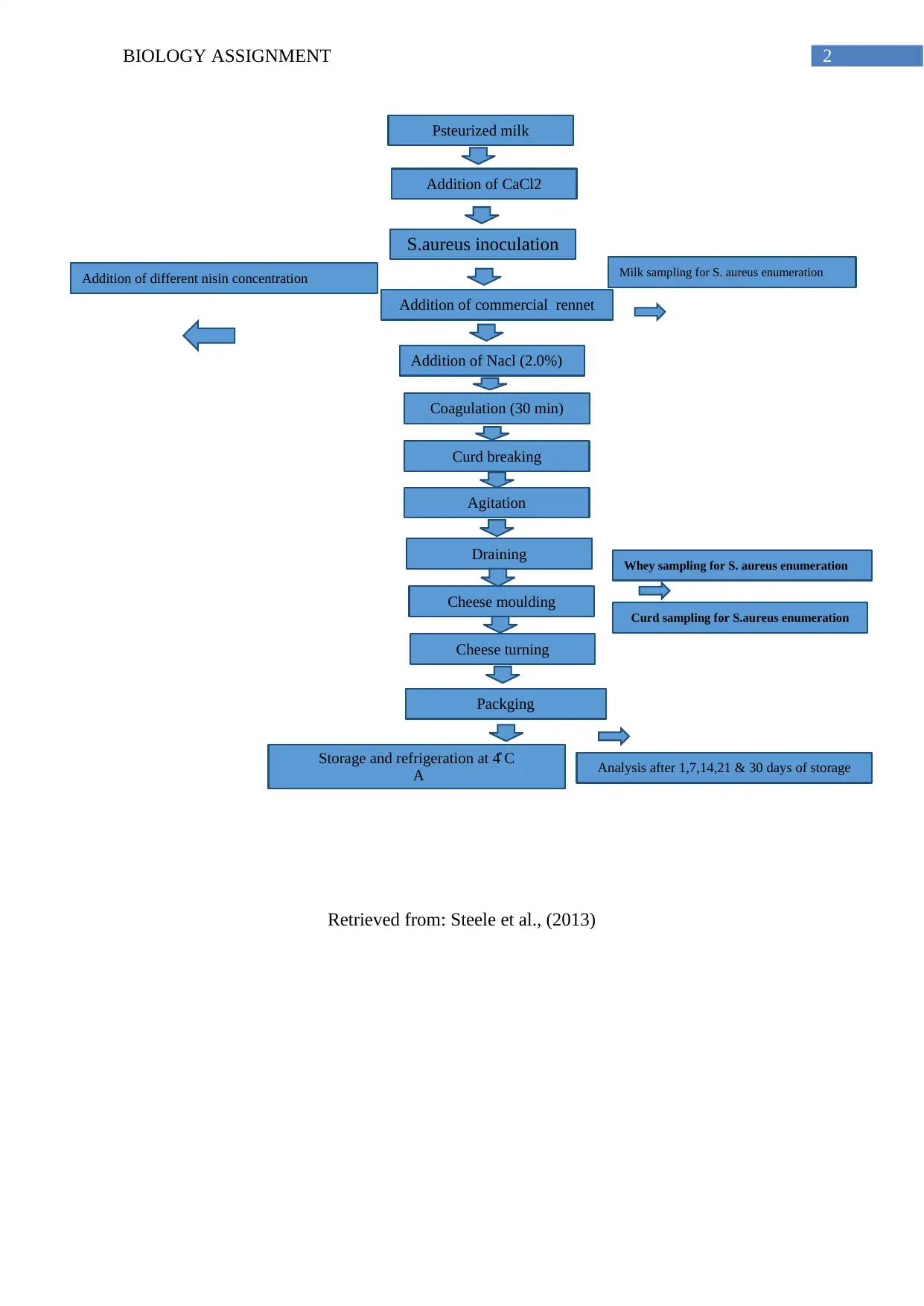
2BIOLOGY ASSIGNMENT
Psteurized milk
S.aureus inoculation
Addition of commercial rennet
Addition of Nacl (2.0%)
Coagulation (30 min)
Curd breaking
Agitation
Draining
Cheese moulding
Cheese turning
Packging
Storage and refrigeration at 4̊ C
A
Addition of CaCl2
Milk sampling for S. aureus enumeration
Addition of different nisin concentration
Whey sampling for S. aureus enumeration
Curd sampling for S.aureus enumeration
Analysis after 1,7,14,21 & 30 days of storage
Retrieved from: Steele et al., (2013)
Psteurized milk
S.aureus inoculation
Addition of commercial rennet
Addition of Nacl (2.0%)
Coagulation (30 min)
Curd breaking
Agitation
Draining
Cheese moulding
Cheese turning
Packging
Storage and refrigeration at 4̊ C
A
Addition of CaCl2
Milk sampling for S. aureus enumeration
Addition of different nisin concentration
Whey sampling for S. aureus enumeration
Curd sampling for S.aureus enumeration
Analysis after 1,7,14,21 & 30 days of storage
Retrieved from: Steele et al., (2013)
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
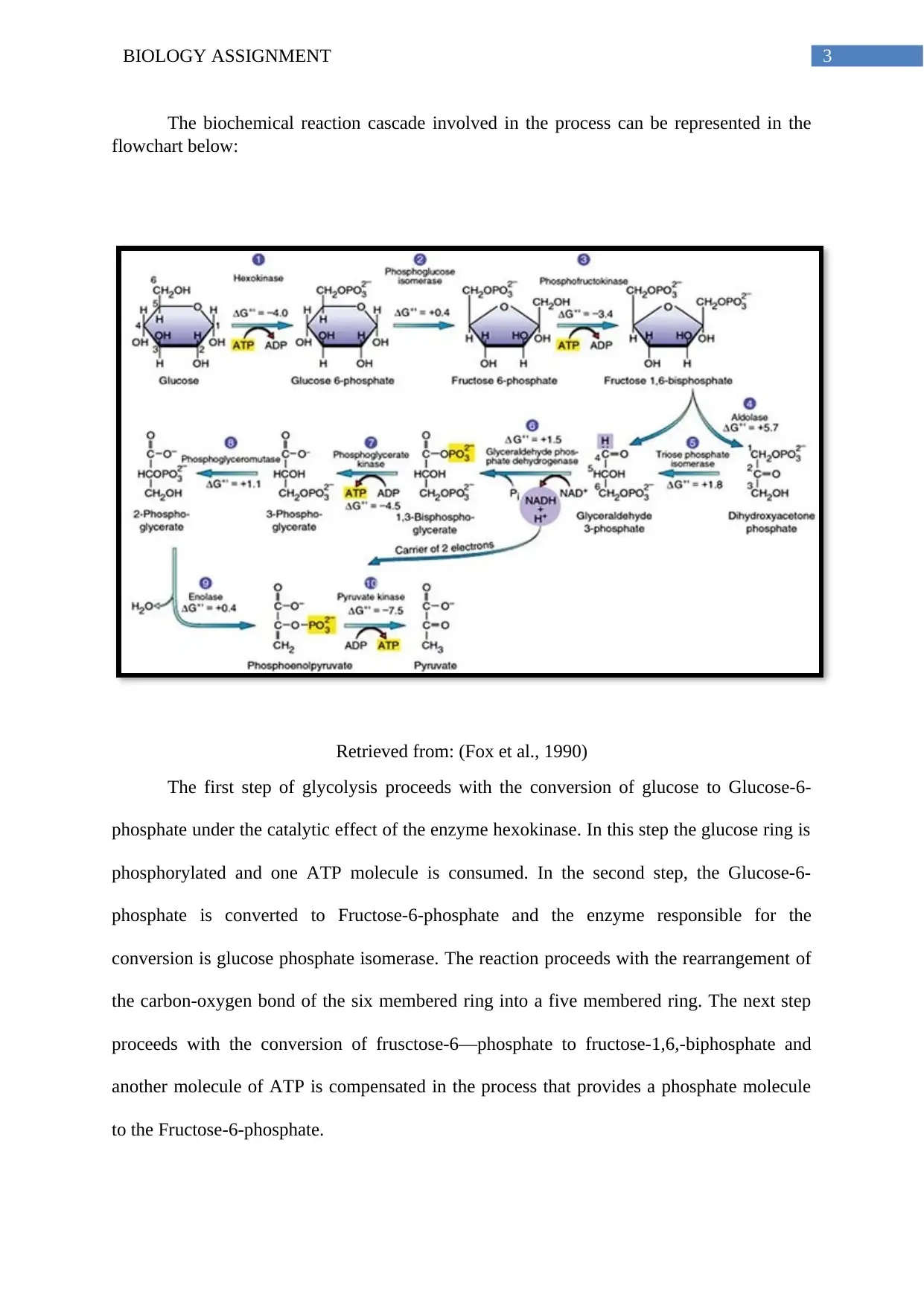
3BIOLOGY ASSIGNMENT
The biochemical reaction cascade involved in the process can be represented in the
flowchart below:
Retrieved from: (Fox et al., 1990)
The first step of glycolysis proceeds with the conversion of glucose to Glucose-6-
phosphate under the catalytic effect of the enzyme hexokinase. In this step the glucose ring is
phosphorylated and one ATP molecule is consumed. In the second step, the Glucose-6-
phosphate is converted to Fructose-6-phosphate and the enzyme responsible for the
conversion is glucose phosphate isomerase. The reaction proceeds with the rearrangement of
the carbon-oxygen bond of the six membered ring into a five membered ring. The next step
proceeds with the conversion of frusctose-6—phosphate to fructose-1,6,-biphosphate and
another molecule of ATP is compensated in the process that provides a phosphate molecule
to the Fructose-6-phosphate.
The biochemical reaction cascade involved in the process can be represented in the
flowchart below:
Retrieved from: (Fox et al., 1990)
The first step of glycolysis proceeds with the conversion of glucose to Glucose-6-
phosphate under the catalytic effect of the enzyme hexokinase. In this step the glucose ring is
phosphorylated and one ATP molecule is consumed. In the second step, the Glucose-6-
phosphate is converted to Fructose-6-phosphate and the enzyme responsible for the
conversion is glucose phosphate isomerase. The reaction proceeds with the rearrangement of
the carbon-oxygen bond of the six membered ring into a five membered ring. The next step
proceeds with the conversion of frusctose-6—phosphate to fructose-1,6,-biphosphate and
another molecule of ATP is compensated in the process that provides a phosphate molecule
to the Fructose-6-phosphate.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

4BIOLOGY ASSIGNMENT
The next step proceeds with the conversion of Fructose-1,6-biphosphate to two sugars
namely, DHAP and GAP. Further, the enzyme triphosphate isomerase interconvert both the
molecules. In the next step, GADPH enzyme dehydrogenates and adds a phosphate group to
form 1,3-biphosphoglycerate. Also, in the next step phosphoglycerate kinase transfers a
phosphate group from 1,3-biphosphoglycerate to form ADP that forms ATP and 3-
phosphoglycerate. In the next step, phosphoglycerate mutase transfers the phosphate group
from 3-phosphoglycerate to form 2-phosphoglycerate. Further, the enzyme Enolase removes
one water molecule from 2-phosphoglycerate to form phosphophenolpyruvic acid. Finally,
the pyruvate enzyme converts phosphophenol pyruvate to pyruvate.
The ATP generated can be explained as:
Steps 1 and 3= subtraction of 2ATP
Steps 7 and 10= addition of 4 ATP
Net ATP produced= 2 ATP
The next step proceeds with the conversion of Fructose-1,6-biphosphate to two sugars
namely, DHAP and GAP. Further, the enzyme triphosphate isomerase interconvert both the
molecules. In the next step, GADPH enzyme dehydrogenates and adds a phosphate group to
form 1,3-biphosphoglycerate. Also, in the next step phosphoglycerate kinase transfers a
phosphate group from 1,3-biphosphoglycerate to form ADP that forms ATP and 3-
phosphoglycerate. In the next step, phosphoglycerate mutase transfers the phosphate group
from 3-phosphoglycerate to form 2-phosphoglycerate. Further, the enzyme Enolase removes
one water molecule from 2-phosphoglycerate to form phosphophenolpyruvic acid. Finally,
the pyruvate enzyme converts phosphophenol pyruvate to pyruvate.
The ATP generated can be explained as:
Steps 1 and 3= subtraction of 2ATP
Steps 7 and 10= addition of 4 ATP
Net ATP produced= 2 ATP

5BIOLOGY ASSIGNMENT
References:
Fox, P. F., Lucey, J. A., & Cogan, T. M. (1990). Glycolysis and related reactions during
cheese manufacture and ripening. Critical Reviews in Food Science &
Nutrition, 29(4), 237-253.
Steele, J., Broadbent, J., & Kok, J. (2013). Perspectives on the contribution of lactic acid
bacteria to cheese flavor development. Current opinion in biotechnology, 24(2), 135-
141.
Linares, D. M., del Río, B., Ladero, V., Redruello, B., Martín, M. C., Fernández, M., &
Alvarez, M. A. (2013). The putrescine biosynthesis pathway in Lactococcus lactis is
transcriptionally regulated by carbon catabolic repression, mediated by
CcpA. International journal of food microbiology, 165(1), 43-50.
References:
Fox, P. F., Lucey, J. A., & Cogan, T. M. (1990). Glycolysis and related reactions during
cheese manufacture and ripening. Critical Reviews in Food Science &
Nutrition, 29(4), 237-253.
Steele, J., Broadbent, J., & Kok, J. (2013). Perspectives on the contribution of lactic acid
bacteria to cheese flavor development. Current opinion in biotechnology, 24(2), 135-
141.
Linares, D. M., del Río, B., Ladero, V., Redruello, B., Martín, M. C., Fernández, M., &
Alvarez, M. A. (2013). The putrescine biosynthesis pathway in Lactococcus lactis is
transcriptionally regulated by carbon catabolic repression, mediated by
CcpA. International journal of food microbiology, 165(1), 43-50.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 6
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.