University Biology: Genome Analysis Homework Solution
VerifiedAdded on 2021/04/24
|7
|1372
|39
Homework Assignment
AI Summary
This document provides a detailed solution to a genome analysis homework assignment. The solution addresses several key concepts in molecular biology and genomics. The first question explains horizontal gene transfer, specifically the transfer of genetic material between prokaryotes and eukaryotes. Question two discusses the number of genes identified in Escherichia coli genomes. Question three explores different types of transposable elements like LINE 1 (L1), Alu elements, and SVA elements, and their impact on genetic mutations and diseases. The fourth question describes the Hi-C technique used to determine the spatial organization of chromatin. Finally, question five analyzes a DNA segment, including transcript numbers, exon/intron counts, DNase sensitivity, histone marks, and tandem repeats. The document includes a comprehensive list of cited works.

Surname: 1
Course:
Tutor:
Date:
Genome
1. Question one
Horizontal gene transfer, also known as lateral gene transfer, has been established to occur
between prokaryotes and eukaryotes. This mode of genetic material transfer occurs between the
symbiotic prokaryotes found within eukaryotic cells. The prokaryote’s genome gets inserted into
the eukaryotic (host’s) genome and when successfully transcribed, it confers newer functions in
the eukaryotes. The researcher’s used a common endosymbiont Wolbachia pipientis and
computationally analyzed its host’s genomes for copies of the bacterial genes. Wolbachia
pipientis’ genetic base sequences were identified and cross referenced with those of four insects
and nematode species where entire Wolbachia genome and other short sequences of base pairs
were identified as insertions in the test species being examined. Three other insect that are not
common hosts for the bacteria had their genomes extracted and their sequences examined for the
symbiont’s genome. This provided proof of lateral gene transfers in eukaryotic hosts that do not
possess endosymbionts. The researchers also located specific regions of bacterial genome that
had been transcribed in the eukaryotes and inferred the implication of translation of such genes
(Julie et al, 1753-1756).
2. Question two
Course:
Tutor:
Date:
Genome
1. Question one
Horizontal gene transfer, also known as lateral gene transfer, has been established to occur
between prokaryotes and eukaryotes. This mode of genetic material transfer occurs between the
symbiotic prokaryotes found within eukaryotic cells. The prokaryote’s genome gets inserted into
the eukaryotic (host’s) genome and when successfully transcribed, it confers newer functions in
the eukaryotes. The researcher’s used a common endosymbiont Wolbachia pipientis and
computationally analyzed its host’s genomes for copies of the bacterial genes. Wolbachia
pipientis’ genetic base sequences were identified and cross referenced with those of four insects
and nematode species where entire Wolbachia genome and other short sequences of base pairs
were identified as insertions in the test species being examined. Three other insect that are not
common hosts for the bacteria had their genomes extracted and their sequences examined for the
symbiont’s genome. This provided proof of lateral gene transfers in eukaryotic hosts that do not
possess endosymbionts. The researchers also located specific regions of bacterial genome that
had been transcribed in the eukaryotes and inferred the implication of translation of such genes
(Julie et al, 1753-1756).
2. Question two
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Surname: 2
With newer sequences being added on a daily basis, successful complete Escherichia coli
genomes sequenced add up to a total of 10120 genomes. Among th sequences identified, there
are 94000 non-redundant genes established in Escherichia coli strains with 4000 core and 90000
pan genes.
3. Question three
Long Interspersed Nuclear Element (LINE) 1 (L1)
LINES are classified are class I transposable elements. The L1 is also referred to as clade
LINE-1, and it is a non-Long terminal repeats (LTRs) retro transposon (Richard and Mark, 691-
703). It is 6 kilobases (kb) long and contains a 5’ end untranslating region (UTR) that possess
internal RNA polymerase II promoter activity. Its base sequence also contains two open Reading
Frames (ORFs) each of which encodes a different functional unit (Babushok and Kazazian, 527-
539): ORF1encodes a RNA-binding protein whereas ORF2 encodes a protein with endonuclease
activity (Martin and Jose, 115). L1 also has a 3’ end untranslating region that is made up of
polyadenylation signals ending with an oligo dA-rich tail that varies in length for different
sequences (Babushok and Kazazian, 527-539). This element causes genetic insertion mutation
that result in most inherited diseases. LINE 1 insertion on the human factor VIII causes the
production of un-functional clotting factor VIII that result in hemophilia. Also its insertion in the
APC gene results in the development of colon cancer.
Alu element
This is another typical example of a non-LTRs retro transposon that contains a million
copies in the human genome. They are mostly 300 base pairs in length with two sequences
derived from 7SL RNA gene combining to form dimeric structures separated by A-rich linker
With newer sequences being added on a daily basis, successful complete Escherichia coli
genomes sequenced add up to a total of 10120 genomes. Among th sequences identified, there
are 94000 non-redundant genes established in Escherichia coli strains with 4000 core and 90000
pan genes.
3. Question three
Long Interspersed Nuclear Element (LINE) 1 (L1)
LINES are classified are class I transposable elements. The L1 is also referred to as clade
LINE-1, and it is a non-Long terminal repeats (LTRs) retro transposon (Richard and Mark, 691-
703). It is 6 kilobases (kb) long and contains a 5’ end untranslating region (UTR) that possess
internal RNA polymerase II promoter activity. Its base sequence also contains two open Reading
Frames (ORFs) each of which encodes a different functional unit (Babushok and Kazazian, 527-
539): ORF1encodes a RNA-binding protein whereas ORF2 encodes a protein with endonuclease
activity (Martin and Jose, 115). L1 also has a 3’ end untranslating region that is made up of
polyadenylation signals ending with an oligo dA-rich tail that varies in length for different
sequences (Babushok and Kazazian, 527-539). This element causes genetic insertion mutation
that result in most inherited diseases. LINE 1 insertion on the human factor VIII causes the
production of un-functional clotting factor VIII that result in hemophilia. Also its insertion in the
APC gene results in the development of colon cancer.
Alu element
This is another typical example of a non-LTRs retro transposon that contains a million
copies in the human genome. They are mostly 300 base pairs in length with two sequences
derived from 7SL RNA gene combining to form dimeric structures separated by A-rich linker

Surname: 3
regions (Kreigs et al, 158-61). They are similar in structure to the 3’ end of L1, however, their 5’
end has an internal RNA polymerase III promoter activity. It is also responsible for generating
insertion mutations that alter gene expression of the affected gene. Insertion of Alu element on
the human ALMS1 gene at exon 8, 10 and 16, results in Alström syndrome, a genetic disorder
that is characterized by blindness and obesity. This is so as the gene encodes a protein
responsible for insulin resistance, hypogonadism and heart disease (Songmi et al, 70-77).
SVA elements
Also a non-LTRs retro transposon with up to 3000 copies in the human genome, SVA
elements are often 2 kilobases long with various hexamer repeat region, Alu-like regions, a
number of tandem repeats and HERV-K10-like region with the common 3’ polyadenylation
signals that is typical of non-LTRs retro transposons (Wang et al, 994-1007). Also responsible
for genetic insertion mutation, its nsertion into the α-spectin gene causes the inheritable familial
elliptocytosis disorder (Eric et al, 1444-1445).
4. Question 4
Hi-C is a typical example of the chromosomal conformation technologies (3C
technologies) that are used in molecular biology to determine the spatial organization of
chromatin within a cell. This technique uses high-throughput sequencing (Hakim, 1068) to
simultaneously quantify the number of interactions of genomic loci between all possible pairs of
fragments. This thus provides a count of all the possible pairwise interactions between complete
genomes. The method involves crosslinking of chromatin with formaldehyde followed by the
digestion and re-ligation with biotin-labeled nucleotide at the junction between DNA fragments
that are covalently bound to each other. The ligation products are sequences to produce heatmaps
regions (Kreigs et al, 158-61). They are similar in structure to the 3’ end of L1, however, their 5’
end has an internal RNA polymerase III promoter activity. It is also responsible for generating
insertion mutations that alter gene expression of the affected gene. Insertion of Alu element on
the human ALMS1 gene at exon 8, 10 and 16, results in Alström syndrome, a genetic disorder
that is characterized by blindness and obesity. This is so as the gene encodes a protein
responsible for insulin resistance, hypogonadism and heart disease (Songmi et al, 70-77).
SVA elements
Also a non-LTRs retro transposon with up to 3000 copies in the human genome, SVA
elements are often 2 kilobases long with various hexamer repeat region, Alu-like regions, a
number of tandem repeats and HERV-K10-like region with the common 3’ polyadenylation
signals that is typical of non-LTRs retro transposons (Wang et al, 994-1007). Also responsible
for genetic insertion mutation, its nsertion into the α-spectin gene causes the inheritable familial
elliptocytosis disorder (Eric et al, 1444-1445).
4. Question 4
Hi-C is a typical example of the chromosomal conformation technologies (3C
technologies) that are used in molecular biology to determine the spatial organization of
chromatin within a cell. This technique uses high-throughput sequencing (Hakim, 1068) to
simultaneously quantify the number of interactions of genomic loci between all possible pairs of
fragments. This thus provides a count of all the possible pairwise interactions between complete
genomes. The method involves crosslinking of chromatin with formaldehyde followed by the
digestion and re-ligation with biotin-labeled nucleotide at the junction between DNA fragments
that are covalently bound to each other. The ligation products are sequences to produce heatmaps
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Surname: 4
that identify Hi-C interactions as areas marked by dark red color (Belton et al, 268-276). The
peaks of the red “triangles” indicate the first covalent interactions between sequences, that is, the
initial point Hi-C interactions.
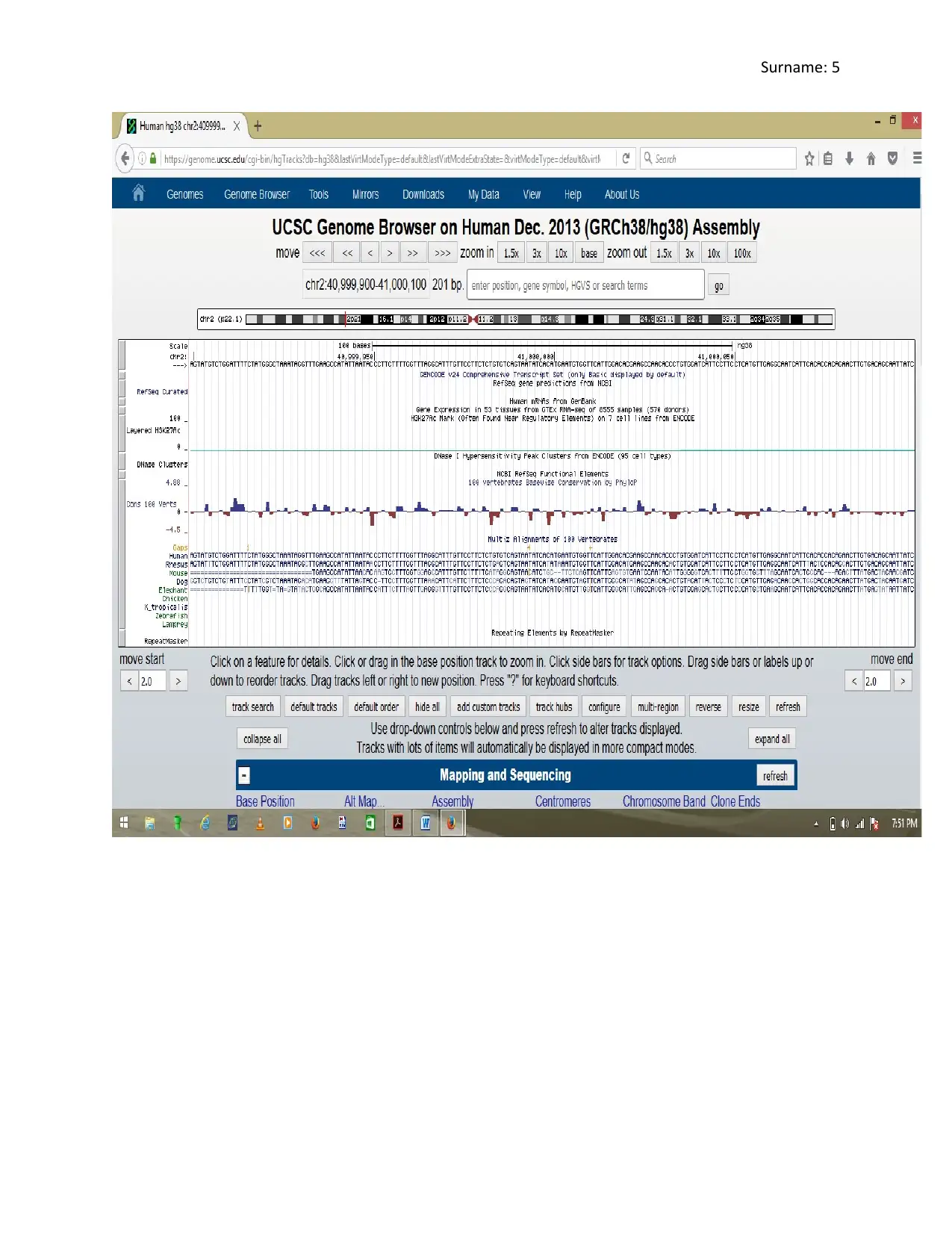
5. Question Five
The segment outlines 24 comprehensive transcripts, hence 24 mRNAs. There are
approximately 40 exons and 37 introns in the identified base sequence. The DNase 1
hypersensitivity for this segment is 0 meaning it cannot be spliced by DNase and hence, it is a
stable region of the chromosome. The position of histone marks can be identified at point 1 and 4
within the DNA segment. The bound histone protein helps in protecting the DNA segment from
attacks by endonucleases. This works to enhance the stability of the DNA segment. The DNA
segment contains many tandem repeats of bases AAA and TTT with many highly conserved
areas.
that identify Hi-C interactions as areas marked by dark red color (Belton et al, 268-276). The
peaks of the red “triangles” indicate the first covalent interactions between sequences, that is, the
initial point Hi-C interactions.
5. Question Five
The segment outlines 24 comprehensive transcripts, hence 24 mRNAs. There are
approximately 40 exons and 37 introns in the identified base sequence. The DNase 1
hypersensitivity for this segment is 0 meaning it cannot be spliced by DNase and hence, it is a
stable region of the chromosome. The position of histone marks can be identified at point 1 and 4
within the DNA segment. The bound histone protein helps in protecting the DNA segment from
attacks by endonucleases. This works to enhance the stability of the DNA segment. The DNA
segment contains many tandem repeats of bases AAA and TTT with many highly conserved
areas.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
Surname: 5

Surname: 6
Work cited
Babushok D. V. and Kazazian H. H. Jr. Progress in Understanding the Biology of the
Human Mutagen LINE-1. Hum Mutat. 2007; no 28: 527-539
Belton M., McCord P., Gibcus J. H., Naumova N., Zhan Y. and Dekker J. Hi-C: A
Comprehensive Technique to Capture the Conformation of Genomes. Methods, 2012 Nov;
Volume 58, no 3: 268-276
Eric M. Ostertag, John L. Goodier, Yue Zhang and Haig H. Kazazian Jr. SVA Elements
are Nonautonomous Retro transposons that Cause Disease, Am J Hum Gen, Volume 73, no 6:
1444-14511
Julie C. Dunning Hotopp, Michael E. Clark, Deodoro C. S. G. Oliveira, Jeremy M.
Foster, Peter Fischer and Monica C. Widespread Lateral Gene Transfer from Intracellular
Bacteria to Multicellular Eukaryotes. Science 21 Sep 2007. Volume. 317, no. 5845, pp. 1753-
1756
Hakim Ofir. “Snapshot: Chromosome Confirmation Capture. Cell, Volume 148, no 5:
1068.e1-2
Kreigs J.O., Churakov G., Jurka J., Brosius J. and Schmitz J. Review Evolutionary
History of 7SL RNA-Derived SINES in Supraprimates. Trends Genet. 2007 Apr; Volume 23, no.
4: 158-61
Martin Munoz-Lopez and Jose L. Garcia-Perez. DNA Transposons: Nature and
Applications in Genomics. Curr Genomics, 2010 Apr; Volume. 11, no. 2: 115-128
Work cited
Babushok D. V. and Kazazian H. H. Jr. Progress in Understanding the Biology of the
Human Mutagen LINE-1. Hum Mutat. 2007; no 28: 527-539
Belton M., McCord P., Gibcus J. H., Naumova N., Zhan Y. and Dekker J. Hi-C: A
Comprehensive Technique to Capture the Conformation of Genomes. Methods, 2012 Nov;
Volume 58, no 3: 268-276
Eric M. Ostertag, John L. Goodier, Yue Zhang and Haig H. Kazazian Jr. SVA Elements
are Nonautonomous Retro transposons that Cause Disease, Am J Hum Gen, Volume 73, no 6:
1444-14511
Julie C. Dunning Hotopp, Michael E. Clark, Deodoro C. S. G. Oliveira, Jeremy M.
Foster, Peter Fischer and Monica C. Widespread Lateral Gene Transfer from Intracellular
Bacteria to Multicellular Eukaryotes. Science 21 Sep 2007. Volume. 317, no. 5845, pp. 1753-
1756
Hakim Ofir. “Snapshot: Chromosome Confirmation Capture. Cell, Volume 148, no 5:
1068.e1-2
Kreigs J.O., Churakov G., Jurka J., Brosius J. and Schmitz J. Review Evolutionary
History of 7SL RNA-Derived SINES in Supraprimates. Trends Genet. 2007 Apr; Volume 23, no.
4: 158-61
Martin Munoz-Lopez and Jose L. Garcia-Perez. DNA Transposons: Nature and
Applications in Genomics. Curr Genomics, 2010 Apr; Volume. 11, no. 2: 115-128
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Surname: 7
Songmi Kim, Chun-Sung Cho, Kyudong Han and Jungnam Lee. Structural Variations of
Alu Element and Human Disease. Genomics Inform, 2016 Sep; Volume 14, no 3: 70-77
Richard Cordaux and Mark A. Batzer. The Impact of Retro transposons on Human
Genome Evolution. Nat Rev Genet. Oct 2009. Volume. 10, no. 10: 691-703
Wang H., Xing J., Grover D., Hedges D. J., Han K., Walker J. A. and Betzer M. A. SVA
Elements: A Hominid-Specific Retroposon Family. J Mol Biol. 2005 Dec 9; Volume 354, no 4:
9994-1007
Songmi Kim, Chun-Sung Cho, Kyudong Han and Jungnam Lee. Structural Variations of
Alu Element and Human Disease. Genomics Inform, 2016 Sep; Volume 14, no 3: 70-77
Richard Cordaux and Mark A. Batzer. The Impact of Retro transposons on Human
Genome Evolution. Nat Rev Genet. Oct 2009. Volume. 10, no. 10: 691-703
Wang H., Xing J., Grover D., Hedges D. J., Han K., Walker J. A. and Betzer M. A. SVA
Elements: A Hominid-Specific Retroposon Family. J Mol Biol. 2005 Dec 9; Volume 354, no 4:
9994-1007
1 out of 7
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.