Molecular Biology 300817: PCR Lab Report, Site-directed mutagenesis
VerifiedAdded on 2022/08/20
|7
|890
|17
Report
AI Summary
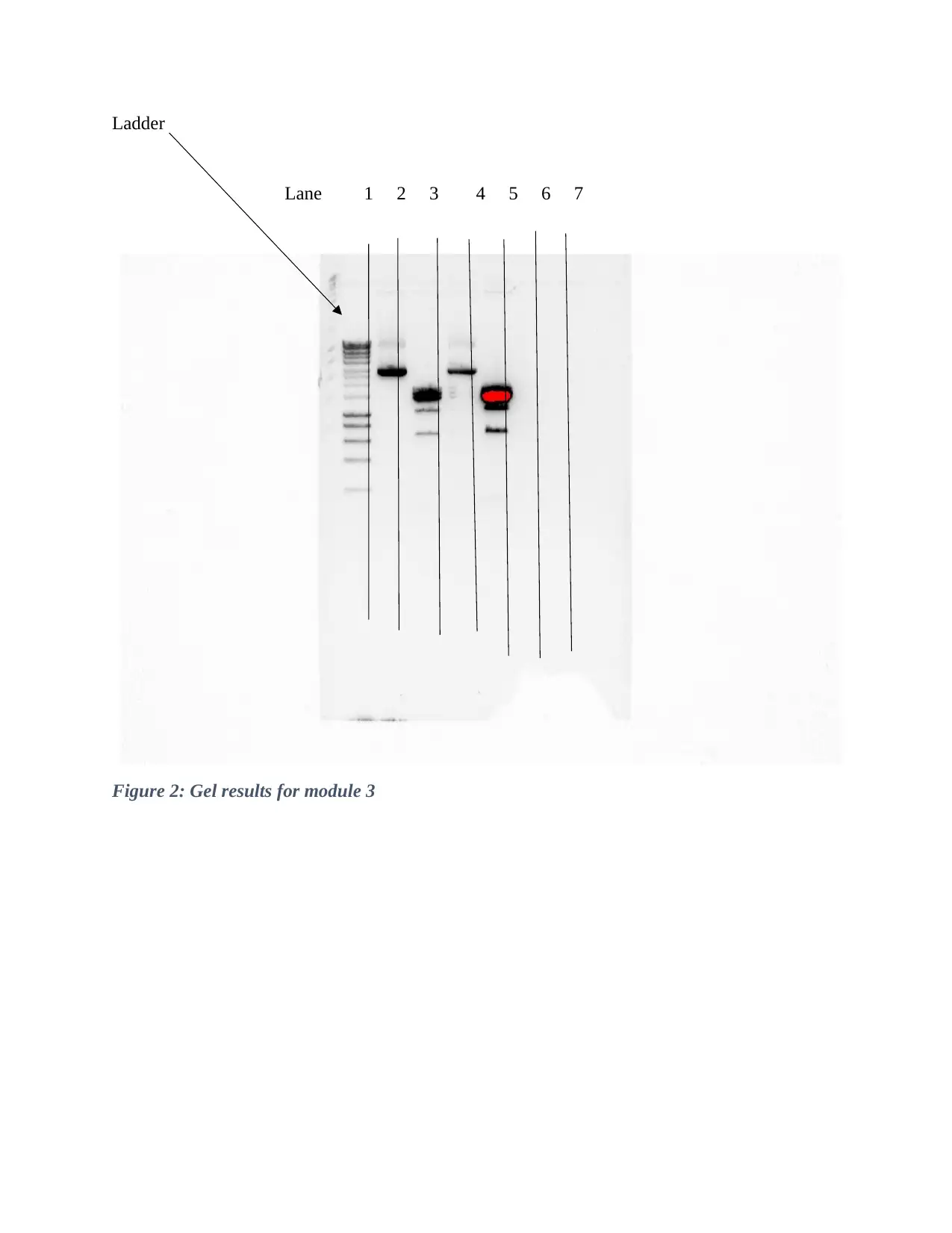
This PCR lab report details an experiment focused on introducing an R204Q point mutation into TP63 cDNA through PCR reactions. The experiment involved testing primer pairs at different annealing temperatures, ligating the purified PCR amplicon into a plasmid, and transforming the result into competent E. coli for cloning. Furthermore, it aimed to digest the purified plasmid using a restriction endonuclease enzyme. The report outlines the procedures for each module, including gel electrophoresis and dilution techniques, along with the results obtained. Despite some initial issues with the results in module 2, the experiment proceeded to successfully synthesize amplicons, determine optimal concentrations, and analyze the size of the amplicons and plasmids. The report concludes that, despite a gel breaking, the objectives of the experiment were successfully met, demonstrating the successful implementation of site-directed mutagenesis and cloning techniques.
1 out of 7










![[object Object]](/_next/static/media/star-bottom.7253800d.svg)