Ligand Discovery Using Competition Dialysis: A Comprehensive Review
VerifiedAdded on 2023/06/08
|5
|811
|139
Report
AI Summary
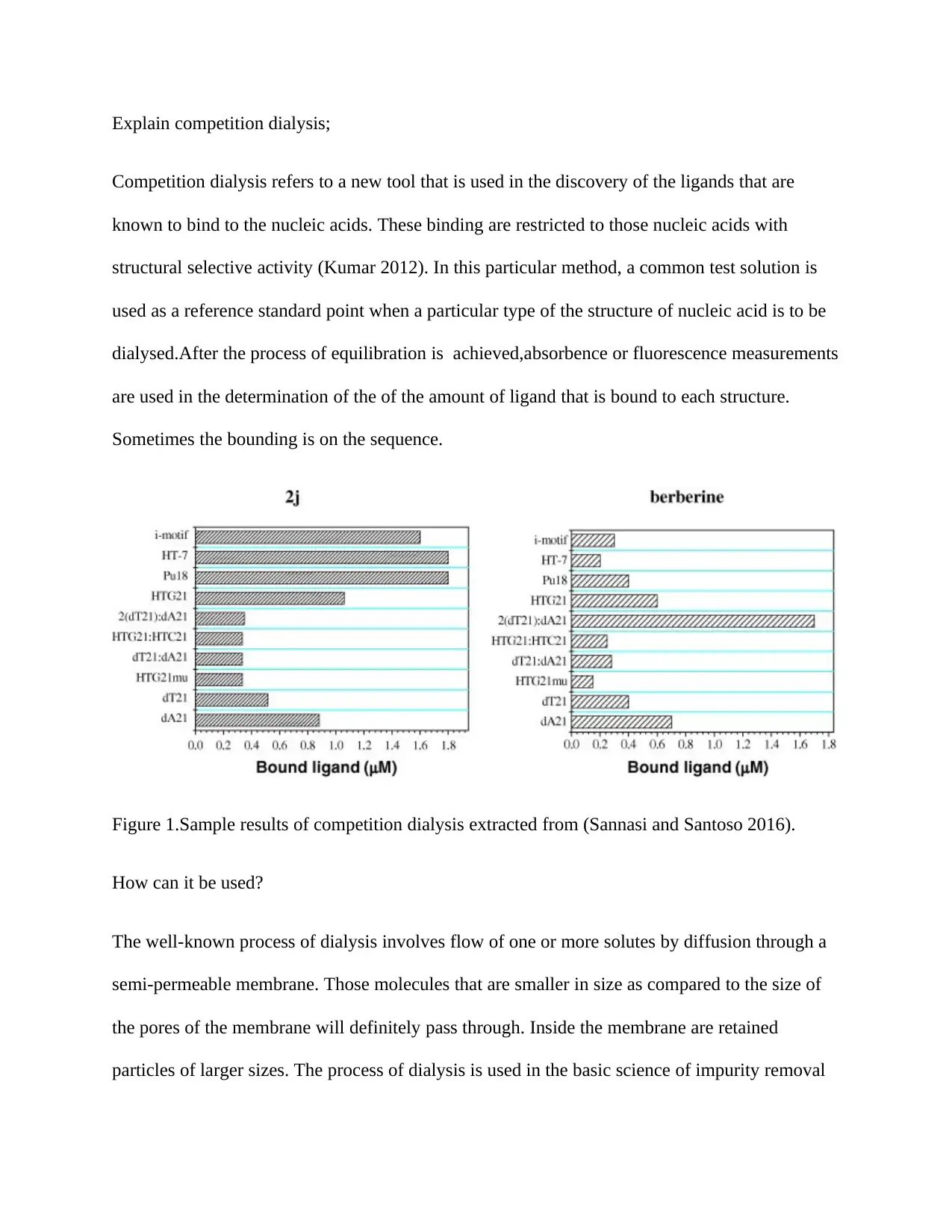
This report discusses competition dialysis, a method used to discover ligands that bind to nucleic acids with structural selectivity. It explains the principles of dialysis, where solutes diffuse through a semi-permeable membrane, and how this process is applied to identify ligands with preferential binding to specific nucleic acid structures. The report also covers the types of ligands suitable for competition dialysis, emphasizing the importance of a convenient spectroscopic monitor for determining ligand concentrations. Specific examples, such as the use of Dispo Dialyzer units, are provided, along with a discussion of different ligands and their affinities to single-stranded DNA and RNA. The report references key studies, including the Chaires review, which highlights the limitations and potential illusions in determining affinity with other nucleic acid structures like double-stranded DNA and RNA. Desklib offers a variety of study tools and resources, including past papers and solved assignments, to further assist students in understanding complex scientific methodologies.
1 out of 5







![[object Object]](/_next/static/media/star-bottom.7253800d.svg)