Detailed Report: DNA Sequence Variation Analysis of Protein P19801-2
VerifiedAdded on 2023/04/20
|26
|3466
|366
Report
AI Summary
This report presents an analysis of DNA sequence variations in protein P19801-2, conducted through BLAST alignment to identify potential isoforms and variations. The analysis includes matching the identified variations with records in dbSNP and connecting them to functional analyses reported in existing literature. The study explores variations in genes like CASP9 and provides graphical representations of these variations across the genome. Mathematical and statistical analyses, including covariance calculations, are applied to understand structural classifications and deviations within the protein's genetic components. The report aims to provide insights that facilitate decision-making and enhance the speed and productivity of research related to protein P19801-2.

BIOINFORMATICS
Name of Institution:
Name of Student:
1
Name of Institution:
Name of Student:
1
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Contents
Introduction......................................................................................................................................3
Literature..........................................................................................................................................4
Fundamental bioinformatics principles,scope and aim...................................................................5
Scope................................................................................................................................................5
Goal..................................................................................................................................................5
The BLAST format of the gene :.....................................................................................................6
The alignment of these potential isoforms with the given proteins as follows:..............................6
The variation of the protein P19801-2 for the gene CASP9..........................................................10
Graphical representation of variation in genes and the regions of genome..................................11
Explanation on the above graphs...................................................................................................15
The helical stretching curves that represents the above graphs.....................................................15
Mathematical analysis of the genes sequences..............................................................................16
The use of covariance in structure classification:..........................................................................16
Relevance of theoretical, mathematical and statistical analisis in genes and regions of genome. 18
Result.............................................................................................................................................19
Discussion......................................................................................................................................19
References......................................................................................................................................20
2
Introduction......................................................................................................................................3
Literature..........................................................................................................................................4
Fundamental bioinformatics principles,scope and aim...................................................................5
Scope................................................................................................................................................5
Goal..................................................................................................................................................5
The BLAST format of the gene :.....................................................................................................6
The alignment of these potential isoforms with the given proteins as follows:..............................6
The variation of the protein P19801-2 for the gene CASP9..........................................................10
Graphical representation of variation in genes and the regions of genome..................................11
Explanation on the above graphs...................................................................................................15
The helical stretching curves that represents the above graphs.....................................................15
Mathematical analysis of the genes sequences..............................................................................16
The use of covariance in structure classification:..........................................................................16
Relevance of theoretical, mathematical and statistical analisis in genes and regions of genome. 18
Result.............................................................................................................................................19
Discussion......................................................................................................................................19
References......................................................................................................................................20
2

Introduction
The nucleotide can be used for the study the variation between the two species. DNA differs
from one organism the variation shall help in understanding the unique nature of the protein.
And also to analyze the evolution of the Genes (Samish, et al., 2015). The variation of this DNA
sequence in a different molecular organism will help understand the molecular structure. The
sequence in the DNA will help in understanding the differences in the protein structure.
The method that could be used to determine the variation in the DNA an organism is protein
structure. The exploration of two species of organism are closely related by evolution ought to
3
The nucleotide can be used for the study the variation between the two species. DNA differs
from one organism the variation shall help in understanding the unique nature of the protein.
And also to analyze the evolution of the Genes (Samish, et al., 2015). The variation of this DNA
sequence in a different molecular organism will help understand the molecular structure. The
sequence in the DNA will help in understanding the differences in the protein structure.
The method that could be used to determine the variation in the DNA an organism is protein
structure. The exploration of two species of organism are closely related by evolution ought to
3
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

have been the difference in their protein sequence (Peng, et al., 2012), for example, the sequence
in the Chimpanzee and humans. Another way of determining the variation is through, DNA
hybridization, “which the process is of hybridizes the genetic information from two different
organisms to ascertain the similarities between them (Moore, et al., 2010). A scientist separates
the strands between them from the two species using heat, which breaks the bonds between the
base pairs that link the two sides of the double helix” (Peng, et al., 2012). Then it is chopped to
small parts which are then mixed up to exhibit generic similarity.
It could be possible to determine the variation through gene sequencing, which could also a huge
chunk of genetic information which is used to compare one gene to another (Marz, et al., 2014).
When it is done this way, it looks the strands of DNA OR RNA or proteins. That exhibit
homogeneous sequence or sequence with similar genes in them. When the evolution is closer
there is the possibility of a closer relationship; the less the similar gene structure has changed in
that period (Moore, et al., 2010).
Variation can also be certain by the use of mitochondrial DNA. Mitochondria are used to assist
in constructing evolution relationships on humans; (Karnovsky, et al., 2012) The DNA of
Mitochondria which contains DNA rather than taking the information from another human
being. But because it is inherited, it exclusively comes from the mother rather than the father.
However, the scientist can examine the DNA on humans (Jelizarow, et al., 2010). To reconstruct
the genetical history of where we came from. However, this method was not perfect according to
Action Science, which has criticized the fact that mitochondrial DNA has a high mutation rate
and could not be accurate add (Fernald, et al., 2011).
4
in the Chimpanzee and humans. Another way of determining the variation is through, DNA
hybridization, “which the process is of hybridizes the genetic information from two different
organisms to ascertain the similarities between them (Moore, et al., 2010). A scientist separates
the strands between them from the two species using heat, which breaks the bonds between the
base pairs that link the two sides of the double helix” (Peng, et al., 2012). Then it is chopped to
small parts which are then mixed up to exhibit generic similarity.
It could be possible to determine the variation through gene sequencing, which could also a huge
chunk of genetic information which is used to compare one gene to another (Marz, et al., 2014).
When it is done this way, it looks the strands of DNA OR RNA or proteins. That exhibit
homogeneous sequence or sequence with similar genes in them. When the evolution is closer
there is the possibility of a closer relationship; the less the similar gene structure has changed in
that period (Moore, et al., 2010).
Variation can also be certain by the use of mitochondrial DNA. Mitochondria are used to assist
in constructing evolution relationships on humans; (Karnovsky, et al., 2012) The DNA of
Mitochondria which contains DNA rather than taking the information from another human
being. But because it is inherited, it exclusively comes from the mother rather than the father.
However, the scientist can examine the DNA on humans (Jelizarow, et al., 2010). To reconstruct
the genetical history of where we came from. However, this method was not perfect according to
Action Science, which has criticized the fact that mitochondrial DNA has a high mutation rate
and could not be accurate add (Fernald, et al., 2011).
4
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Literature
The genetical information gathered from all over the world on the variation. However, the
bioinformatics has been occurring for over fifty hundred years (Troshin, et al., 2011). “The
variation has occurred over the period. Which has actualized the need of the research operation?
The time for the foundation laid was in the 1960s with the publication of computerized to
computerized variation sequence analysis. (Notably, de novo sequence assembly biological
sequence databases and the substitution model). Later on, DNA analysis was critically discussed
due to other disciplines for example (I) molecular biology methods, which could be easily
manipulated to DNA (Tikhvinskiy & Porozov, 2013). As well as sequencing. (ii) Computerized
science, which facilitated the increase in miniaturized and more powerful computers as well as
the novel software which significantly increased bioinformatics information management. The
information (that was generated from the data runs through a blast) manipulated through the
technology to bring clear variation in the protein P1980-2.
The name of the gene in focus continue to be P19801-2, and related information regarding the
protein is derived from UniProt (Suplatov, et al., 2011). BLAST format used for the presentation
of the nucleotide sequence and the BLAST is used for the alignment information “as per
(Fernald, et al., 2011).
Fundamental bioinformatics principles,scope and aim of bioinformatics.
Bioinformatics is a interdisplinary between two displines : computer science and biological
science (Marz, et al., 2014). Difinitions exixts in all over the world on the world wide web of
which some are more inclusive than others. For example bioinformatics being a union of
informatics and biology, could easily be described as the technology that uses computers for
storage, retrival , manupilation , and and fuctional analysis of the gene (Karnovsky, et al., 2012).
5
The genetical information gathered from all over the world on the variation. However, the
bioinformatics has been occurring for over fifty hundred years (Troshin, et al., 2011). “The
variation has occurred over the period. Which has actualized the need of the research operation?
The time for the foundation laid was in the 1960s with the publication of computerized to
computerized variation sequence analysis. (Notably, de novo sequence assembly biological
sequence databases and the substitution model). Later on, DNA analysis was critically discussed
due to other disciplines for example (I) molecular biology methods, which could be easily
manipulated to DNA (Tikhvinskiy & Porozov, 2013). As well as sequencing. (ii) Computerized
science, which facilitated the increase in miniaturized and more powerful computers as well as
the novel software which significantly increased bioinformatics information management. The
information (that was generated from the data runs through a blast) manipulated through the
technology to bring clear variation in the protein P1980-2.
The name of the gene in focus continue to be P19801-2, and related information regarding the
protein is derived from UniProt (Suplatov, et al., 2011). BLAST format used for the presentation
of the nucleotide sequence and the BLAST is used for the alignment information “as per
(Fernald, et al., 2011).
Fundamental bioinformatics principles,scope and aim of bioinformatics.
Bioinformatics is a interdisplinary between two displines : computer science and biological
science (Marz, et al., 2014). Difinitions exixts in all over the world on the world wide web of
which some are more inclusive than others. For example bioinformatics being a union of
informatics and biology, could easily be described as the technology that uses computers for
storage, retrival , manupilation , and and fuctional analysis of the gene (Karnovsky, et al., 2012).
5

Scope
Bioinformatics contains two field which is the development of computation tool of database and
the application of these database to generate biological knowledge to better understanrd the
organisms and their genetical make up (Karnovsky, et al., 2012). The two field work hand in
hand. Writing software are for sequencing, structuring and functioning analysis and also to
develop biological analytical software (Karnovsky, et al., 2012). The new computerized
software could help solve the problem of biological data.
From the three aspects of bioinformatics analysis, the interaction from the database produced
integrated result that is desired in biological research (Marz, et al., 2014).
Refer to
http://www.cambridge.org/9780521840989
Goal
Despite the distinction, bioinformatics helps to understand a living cell, its fuction and
molecular structure. Bioinformatics can generate new insight and provide a global perspective of
the the cell through analyzing raw molecular and structural data.of the cell. The reasons on how
the cell fuction can be easily understood through analyzing the genetical make up for instance
how its DNA is translated to RNA.
6
Bioinformatics contains two field which is the development of computation tool of database and
the application of these database to generate biological knowledge to better understanrd the
organisms and their genetical make up (Karnovsky, et al., 2012). The two field work hand in
hand. Writing software are for sequencing, structuring and functioning analysis and also to
develop biological analytical software (Karnovsky, et al., 2012). The new computerized
software could help solve the problem of biological data.
From the three aspects of bioinformatics analysis, the interaction from the database produced
integrated result that is desired in biological research (Marz, et al., 2014).
Refer to
http://www.cambridge.org/9780521840989
Goal
Despite the distinction, bioinformatics helps to understand a living cell, its fuction and
molecular structure. Bioinformatics can generate new insight and provide a global perspective of
the the cell through analyzing raw molecular and structural data.of the cell. The reasons on how
the cell fuction can be easily understood through analyzing the genetical make up for instance
how its DNA is translated to RNA.
6
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

The BLAST format of the gene :
>sp|p19801-2|aoc1_human isoform 2 of amyloid-sensitive amine oxidase [copper-containing]
os=Homo sapiens ox=9606 Gn=aoc1
mpalgwavaailmlqtamaepspgtlprkagvfsdlsnqelkavhsflwskkelrlqpss
tttmakntvfliemllpkkyhvlrfldkgerhpvrearaviffgdqehpnvtefavgplp
gpcymralsprpgyqsswasrpistaeyallyhtlqeatkplhqfflnttgfsfqdchdr
claftdvaprgvasgqrrswliiqryvegyflhptglellvdhgstdaghwaveqvwyng
kfygspeelarkyadgevdvvvledplpggkghdsteepplfsshkprgdfpspihvsgp
rlvqphgprfrlegnavlyggwsfafrlrsssglqvlnvhfggeriayevsvqeavalyg
ghtpagmqtkyldvgwglgsvthelapgidcpetatfldtfhyydaddpvhypralclfe
mptgvplrrhfnsnfkggfnfyaglkgqvlvlrttstvynydyiwdfifypngvmeakmh
atgyvhatfytpeglrhgtrlhthlignihthlvhyrvdldvagtknsfqtlqmklenit
npwsprhrvvqptleqtqyswerqaafrfkrklpkyllftspqenpwghkrtyrlqihsm
adqvlppgwqeeqaitwarteggqpralsqaaspvpgryplavtkyreselcsssiyhqn
dpwhppvvfeqflhnnenienedlvawvtvgflhiphsedipntatpgnsvgfllrpfnf
fpedpslasrdtvivwprdngpnyvqrwipedrdcsmpppfsyngtyrpv
The variation which associated with the protein P1980-2 are,
The alignment of these potential isoforms with the given proteins as follows:
The proteins which are associated with this organisms are namely Eukaryote (Biosample)
(JH921433.1 MULTIGERMTUBI), Eukaryote (Biosample) (JH921437.1), A
(A0A2J8RK70_PONAB), Ascomycota (JH921451.1) (Karnovsky, et al., 2012).
The alignment of the variation sequence is as follows
7
>sp|p19801-2|aoc1_human isoform 2 of amyloid-sensitive amine oxidase [copper-containing]
os=Homo sapiens ox=9606 Gn=aoc1
mpalgwavaailmlqtamaepspgtlprkagvfsdlsnqelkavhsflwskkelrlqpss
tttmakntvfliemllpkkyhvlrfldkgerhpvrearaviffgdqehpnvtefavgplp
gpcymralsprpgyqsswasrpistaeyallyhtlqeatkplhqfflnttgfsfqdchdr
claftdvaprgvasgqrrswliiqryvegyflhptglellvdhgstdaghwaveqvwyng
kfygspeelarkyadgevdvvvledplpggkghdsteepplfsshkprgdfpspihvsgp
rlvqphgprfrlegnavlyggwsfafrlrsssglqvlnvhfggeriayevsvqeavalyg
ghtpagmqtkyldvgwglgsvthelapgidcpetatfldtfhyydaddpvhypralclfe
mptgvplrrhfnsnfkggfnfyaglkgqvlvlrttstvynydyiwdfifypngvmeakmh
atgyvhatfytpeglrhgtrlhthlignihthlvhyrvdldvagtknsfqtlqmklenit
npwsprhrvvqptleqtqyswerqaafrfkrklpkyllftspqenpwghkrtyrlqihsm
adqvlppgwqeeqaitwarteggqpralsqaaspvpgryplavtkyreselcsssiyhqn
dpwhppvvfeqflhnnenienedlvawvtvgflhiphsedipntatpgnsvgfllrpfnf
fpedpslasrdtvivwprdngpnyvqrwipedrdcsmpppfsyngtyrpv
The variation which associated with the protein P1980-2 are,
The alignment of these potential isoforms with the given proteins as follows:
The proteins which are associated with this organisms are namely Eukaryote (Biosample)
(JH921433.1 MULTIGERMTUBI), Eukaryote (Biosample) (JH921437.1), A
(A0A2J8RK70_PONAB), Ascomycota (JH921451.1) (Karnovsky, et al., 2012).
The alignment of the variation sequence is as follows
7
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

P|P19801-2|AOC1 EUKARYOTA
1 MELPITAKAG QQQRQLDRFC RQYIQVTLEL DYPAEEYLRL DAIQQSIFRR
CFSEDVEYTP
61 PPRYKLRVLK ELVKRIETSI QDWDEEAISD DLMNCLTPLL SMSMPNEATA
AQQKSHVTYT
121 LSLLPRQQDI SPSITLHEAR NMLAAAGTTG LRTWEAGLHL GNYLCTNPHL
VRGKSILELG
181 SGTGFLSILC AKYLKPSHVL ATDGDDDVVA SFSTNFYLNG LQDSSDLNGR
ALKWGHPVTG
241 GEDPHWDPER PVDLVLGADL TYDPRNIPPL VSTFRDLFAL YPDAKILIAA
TVRSQETFAK
301 FPEACRKNDF GFEDIEFGML KSEDQEGPFY SDLAHVQLCV ITRT
************************************************************
P|P19801-2|AOC1_ASCOMYCOTA
1 MQLSEAWRKY LTSRSTTYLS TAATHPKTEK KMSPKQARMK TASPSSATMS
LDDLQLQRSA
61 QNPMSQNISP RLLQDGGPGS SSVSELEKAP SLRRPQKVVT VIVGPEDTKE
TYIIHKGIIC
121 YYSPFFNAAF NGNFAEGETQ TMRLDDVNSE TFGLLVDYLY TQQIDVDPKD
YDGNIIPLAQ
8
1 MELPITAKAG QQQRQLDRFC RQYIQVTLEL DYPAEEYLRL DAIQQSIFRR
CFSEDVEYTP
61 PPRYKLRVLK ELVKRIETSI QDWDEEAISD DLMNCLTPLL SMSMPNEATA
AQQKSHVTYT
121 LSLLPRQQDI SPSITLHEAR NMLAAAGTTG LRTWEAGLHL GNYLCTNPHL
VRGKSILELG
181 SGTGFLSILC AKYLKPSHVL ATDGDDDVVA SFSTNFYLNG LQDSSDLNGR
ALKWGHPVTG
241 GEDPHWDPER PVDLVLGADL TYDPRNIPPL VSTFRDLFAL YPDAKILIAA
TVRSQETFAK
301 FPEACRKNDF GFEDIEFGML KSEDQEGPFY SDLAHVQLCV ITRT
************************************************************
P|P19801-2|AOC1_ASCOMYCOTA
1 MQLSEAWRKY LTSRSTTYLS TAATHPKTEK KMSPKQARMK TASPSSATMS
LDDLQLQRSA
61 QNPMSQNISP RLLQDGGPGS SSVSELEKAP SLRRPQKVVT VIVGPEDTKE
TYIIHKGIIC
121 YYSPFFNAAF NGNFAEGETQ TMRLDDVNSE TFGLLVDYLY TQQIDVDPKD
YDGNIIPLAQ
8

181 LWVIAGRFFM PALQNKIMNE LRTMVEWAEE EGLRKFMHFA YEASVERTPL
KSLATDMMAW
241 MTPAAGLQIW ITKGYLPDGM MADIIMSLKK DHIFGAKPTR KFGVLGRAKE
YYVRVGEEAA
301 APKQEKQGVE KMPTPYFSES CVNIALPPLA LSHIKDNLST SPDMASIIVP
EFVKREQQQL
361 LQAEPEELVT IIVSDGDEEE EYMVHRELIC SCSVYFSYIF NSVIDGSKDN
SVTTLRLEDT
421 DPEIFGLVVR WIYTSDIESA EALSLAKLWM LCAEVHMPVL QNRAMDKIRS
LLRAGVWPGE
481 NLDDIKALVD YAFDANTDRL DRFPLLQKAL VDHFAYLPTG ALDTWMEHLP
ALLLVHLTKS
541 LNSHFNRLPM DLQSWELRKD EQYHVEVLDD RE
************************************************************
P|P19801-2|AOC1_Dermateaceae
1 MELPITAKAG QQQRQLDRFC RQYIQVTLEL DYPAEEYLRL DAIQQSIFRR
CFSEDVEYTP
61 PPRYKLRVLK ELVKRIETSI QDWDEEAISD DLMNCLTPLL SMSMPNEATA
AQQKSHVTYT
121 LSLLPRQQDI SPSITLHEAR NMLAAAGTTG LRTWEAGLHL GNYLCTNPHL
VRGKSILELG
9
KSLATDMMAW
241 MTPAAGLQIW ITKGYLPDGM MADIIMSLKK DHIFGAKPTR KFGVLGRAKE
YYVRVGEEAA
301 APKQEKQGVE KMPTPYFSES CVNIALPPLA LSHIKDNLST SPDMASIIVP
EFVKREQQQL
361 LQAEPEELVT IIVSDGDEEE EYMVHRELIC SCSVYFSYIF NSVIDGSKDN
SVTTLRLEDT
421 DPEIFGLVVR WIYTSDIESA EALSLAKLWM LCAEVHMPVL QNRAMDKIRS
LLRAGVWPGE
481 NLDDIKALVD YAFDANTDRL DRFPLLQKAL VDHFAYLPTG ALDTWMEHLP
ALLLVHLTKS
541 LNSHFNRLPM DLQSWELRKD EQYHVEVLDD RE
************************************************************
P|P19801-2|AOC1_Dermateaceae
1 MELPITAKAG QQQRQLDRFC RQYIQVTLEL DYPAEEYLRL DAIQQSIFRR
CFSEDVEYTP
61 PPRYKLRVLK ELVKRIETSI QDWDEEAISD DLMNCLTPLL SMSMPNEATA
AQQKSHVTYT
121 LSLLPRQQDI SPSITLHEAR NMLAAAGTTG LRTWEAGLHL GNYLCTNPHL
VRGKSILELG
9
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

181 SGTGFLSILC AKYLKPSHVL ATDGDDDVVA SFSTNFYLNG LQDSSDLNGR
ALKWGHPVTG
241 GEDPHWDPER PVDLVLGADL TYDPRNIPPL VSTFRDLFAL YPDAKILIAA
TVRSQETFAK
301 FPEACRKNDF GFEDIEFGML KSEDQEGPFY SDLAHVQLCV ITRT
************************************************************
P|P19801-2|AOC1_Helotiales
ORIGIN
1 MDPPSEEPAT PKKHTRNASS GRGALPRRPT RGPLEVADSP ARPSIKSTPP
PNLRQQQSSQ
61 LSTTASQHAT PRVPSPGPGS NLTASFVTAR TSLSPSRPGS RSKDTSNMST
TSPAIQKDFS
121 FLVRPEIYHP LTLLDVPPPF RAASSEIDPS TSLASLLSSG HFRNAAIKAA
QLLTAPGLDV
181 KDHAAIFSLV YTRLSCLTLC NQTPLAAQEV KALEDLNSGY YRDDLTGAHM
VPWELRVLAV
241 RLQGMGFNDA RRGVMGYYDL AREARSMLNK LKRKRRKEEI GDDAARAELE
GIKVWEARLE
301 ELGVRVASAL VEMEDLEGAA RFLGTLKPET GTRLEIQKAL LWLCIGDVEA
ARKCVLGKGD
10
ALKWGHPVTG
241 GEDPHWDPER PVDLVLGADL TYDPRNIPPL VSTFRDLFAL YPDAKILIAA
TVRSQETFAK
301 FPEACRKNDF GFEDIEFGML KSEDQEGPFY SDLAHVQLCV ITRT
************************************************************
P|P19801-2|AOC1_Helotiales
ORIGIN
1 MDPPSEEPAT PKKHTRNASS GRGALPRRPT RGPLEVADSP ARPSIKSTPP
PNLRQQQSSQ
61 LSTTASQHAT PRVPSPGPGS NLTASFVTAR TSLSPSRPGS RSKDTSNMST
TSPAIQKDFS
121 FLVRPEIYHP LTLLDVPPPF RAASSEIDPS TSLASLLSSG HFRNAAIKAA
QLLTAPGLDV
181 KDHAAIFSLV YTRLSCLTLC NQTPLAAQEV KALEDLNSGY YRDDLTGAHM
VPWELRVLAV
241 RLQGMGFNDA RRGVMGYYDL AREARSMLNK LKRKRRKEEI GDDAARAELE
GIKVWEARLE
301 ELGVRVASAL VEMEDLEGAA RFLGTLKPET GTRLEIQKAL LWLCIGDVEA
ARKCVLGKGD
10
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
361 GHEQKVILAL AHMADSDFAA AVETWRALIA SDAAEDDGGE KAMWMQNLGV
CYLYLGRMDE
421 ARTLLESLIS GAQDLHAFHF HALTFNLCTI YELCTERSRG LKIALAERVA
GMQQQQGDGG
481 GSNGGWEKVN GDFKL
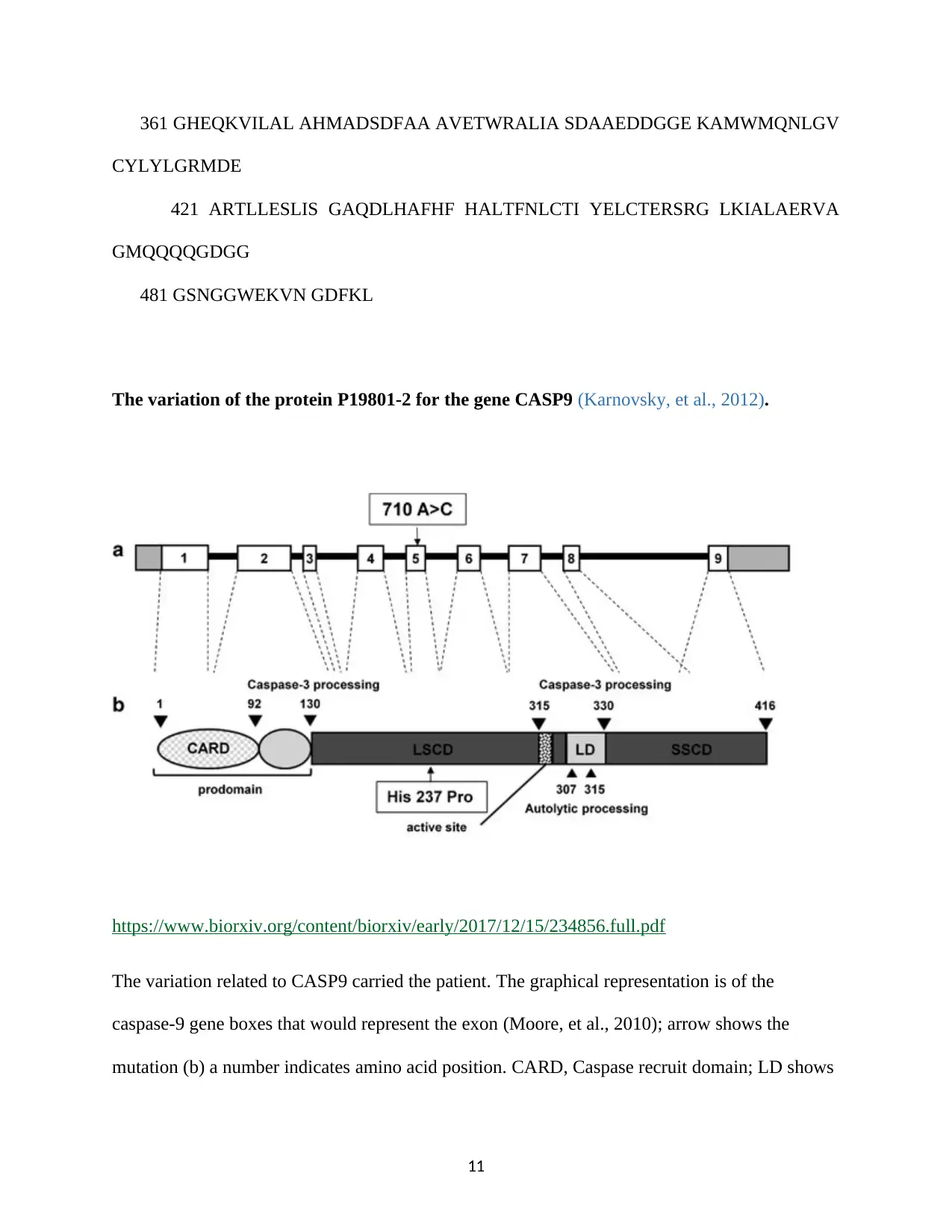
The variation of the protein P19801-2 for the gene CASP9 (Karnovsky, et al., 2012).
https://www.biorxiv.org/content/biorxiv/early/2017/12/15/234856.full.pdf
The variation related to CASP9 carried the patient. The graphical representation is of the
caspase-9 gene boxes that would represent the exon (Moore, et al., 2010); arrow shows the
mutation (b) a number indicates amino acid position. CARD, Caspase recruit domain; LD shows
11
CYLYLGRMDE
421 ARTLLESLIS GAQDLHAFHF HALTFNLCTI YELCTERSRG LKIALAERVA
GMQQQQGDGG
481 GSNGGWEKVN GDFKL
The variation of the protein P19801-2 for the gene CASP9 (Karnovsky, et al., 2012).
https://www.biorxiv.org/content/biorxiv/early/2017/12/15/234856.full.pdf
The variation related to CASP9 carried the patient. The graphical representation is of the
caspase-9 gene boxes that would represent the exon (Moore, et al., 2010); arrow shows the
mutation (b) a number indicates amino acid position. CARD, Caspase recruit domain; LD shows
11
the link between two subunits; LSCD: large subunits catalytic domain; small catalytic domain
(Karnovsky, et al., 2012).
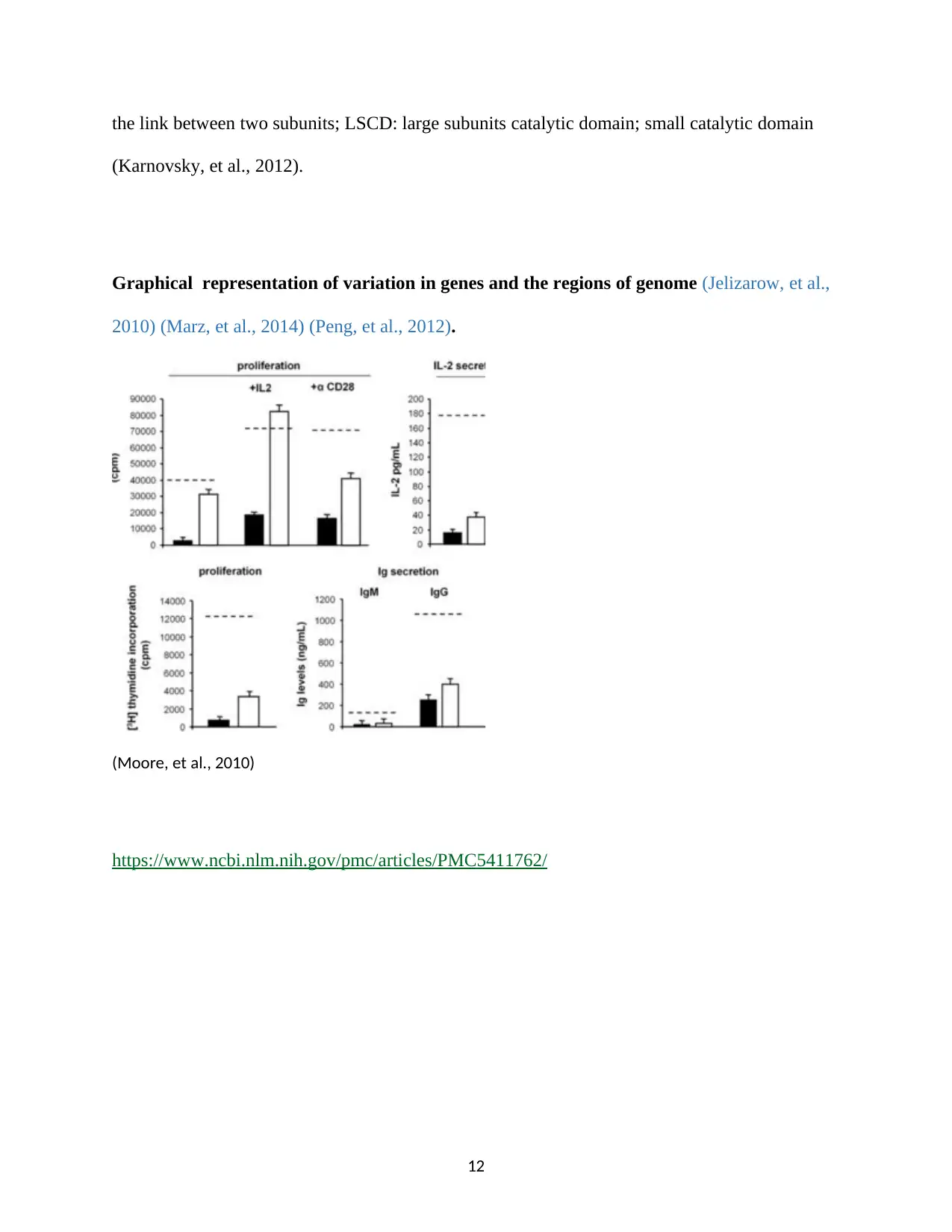
Graphical representation of variation in genes and the regions of genome (Jelizarow, et al.,
2010) (Marz, et al., 2014) (Peng, et al., 2012).
(Moore, et al., 2010)
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5411762/
12
(Karnovsky, et al., 2012).
Graphical representation of variation in genes and the regions of genome (Jelizarow, et al.,
2010) (Marz, et al., 2014) (Peng, et al., 2012).
(Moore, et al., 2010)
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5411762/
12
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 26
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.





