Analysis of PCR, Cloning, and Protein Expression in Biology
VerifiedAdded on 2023/01/17
|5
|755
|29
Practical Assignment
AI Summary
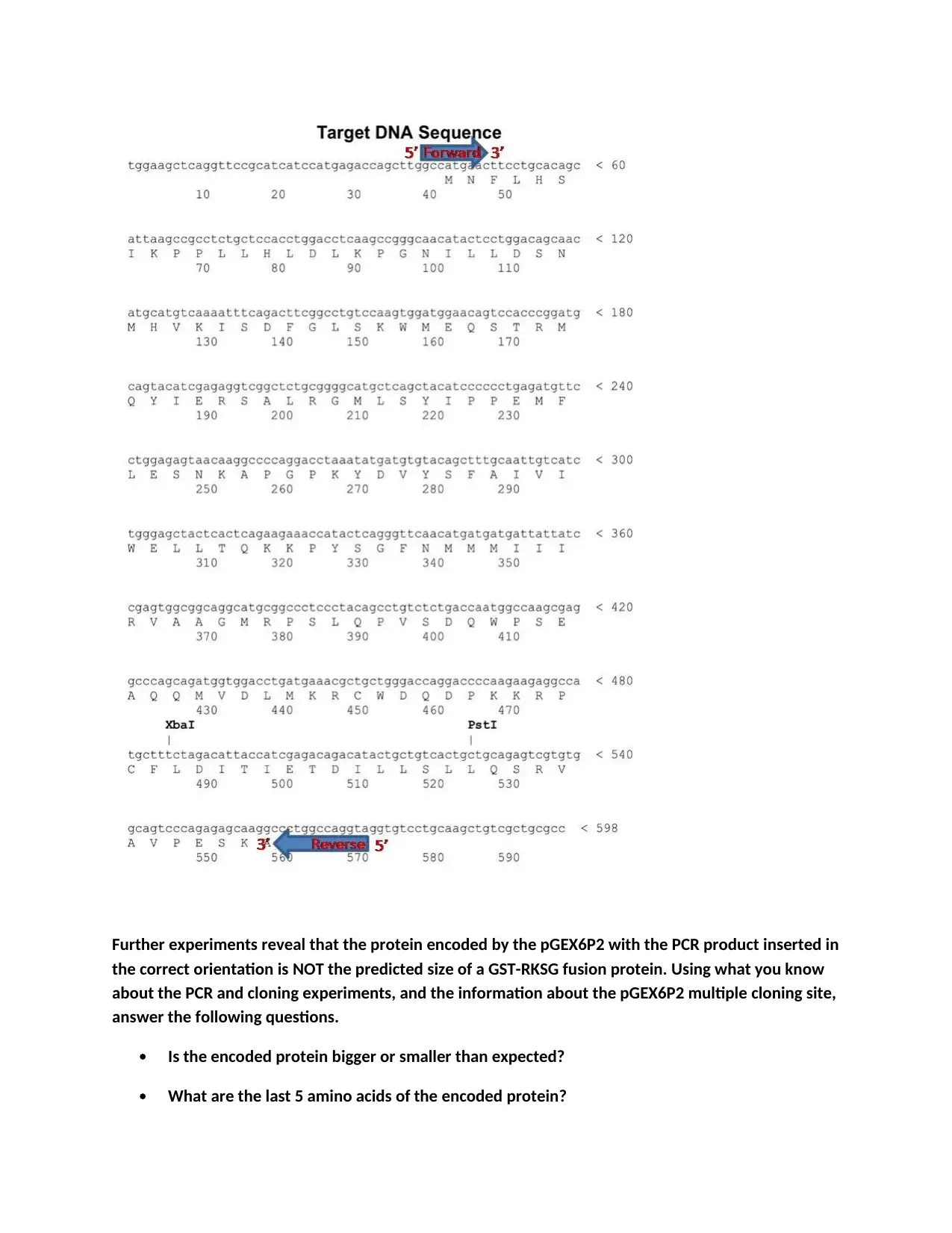
This assignment focuses on analyzing a molecular biology experiment involving PCR amplification, cloning, and protein expression using the pGEX6P2 vector. The first task requires students to interpret and draw a gel electrophoresis image based on the results of restriction enzyme digestions and PCR reactions, labeling bands with their sizes. The second task involves analyzing a target DNA sequence, identifying primer binding sites, and determining the direction of primer extension. Furthermore, students must address discrepancies in the expected protein size, determine the last five amino acids of the protein, and design improved primers to correct errors in the original primer design. The assignment assesses the understanding of plasmid maps, restriction digestion, PCR principles, and protein translation, along with the ability to troubleshoot experimental outcomes and design solutions.
1 out of 5





![[object Object]](/_next/static/media/star-bottom.7253800d.svg)