Plasmid Isolation and Analysis: GeneJet Plasmid Digestion Report
VerifiedAdded on 2022/08/12
|10
|1658
|18
Report
AI Summary
This report details an experiment focused on plasmid DNA isolation, digestion using the HindIII restriction enzyme, and subsequent analysis through restriction mapping. The GeneJet plasmid was isolated using a miniprep protocol involving centrifugation and solutions like sodium hydroxide, TE buffer with RNAse, potassium acetate, and ethanol. Restriction digestion with HindIII was performed, followed by agarose gel electrophoresis to assess digestion. The results, including data from two students, are presented with gel electrophoresis images and a discussion of theoretical concepts such as DNA ladders, the importance of quality kits, chaotropic agents, and the effects of improper experimental procedures. The report also includes explanations of restriction enzyme types (Type IIP and IIS), exonucleases, endonucleases, and the FokI restriction enzyme, along with relevant references and calculations.

Running head: PLASMID ISOLATION. 1
PLASMID ISOLATION.
Student Name
Institution of Affiliation.
PLASMID ISOLATION.
Student Name
Institution of Affiliation.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

PLASMID ISOLATION. 2
Introduction.
This experiment aimed to isolate plasmid DNA, digest it with restriction enzyme HindIII and
thereafter analyze by restriction mapping. The GeneJet plasmid was isolated by a plasmid
miniprep protocol that included a series of centrifugation steps using these solutions Sodium
hydroxide (SDS), TE buffer containing RNAse, potassium acetate, and ethyl alcohol wash
buffer. Restriction digestion of the isolated plasmid occurred in the step where it was put into the
tube containing the HindIII restriction enzyme. To analyze the digested plasmid agarose gel
electrophoresis was done to determine whether digestion took place. From the results obtained
from gel electrophoresis, we can identify the undigested and digested plasmid. The alkaline lysis
method is very important in the field of biotechnology because it’s a faster and cleaner method of
isolating plasmid DNA from cells. This is because of the plasmid DNA characteristic to anneal
after denaturation (Antipov et al., 2016)
RESULTS.
Legends of the well Starting from the left of the gel image:
What is in the sample for each well?
1) 5 uL H2O, 5 uL DNA, 2 uL 6x LD (loading dye)
2) 3.2 uL H2O, 0.2 uL RNase, 5 uL DNA, 1 uL 10 x Buffer, enzyme or HindIII 0.5
uL
3) 3.5 uL H2O, 5 uL DNA, 1 uL 10 x Buffer, enzyme or HindIII 0.5 uL
4) N
Introduction.
This experiment aimed to isolate plasmid DNA, digest it with restriction enzyme HindIII and
thereafter analyze by restriction mapping. The GeneJet plasmid was isolated by a plasmid
miniprep protocol that included a series of centrifugation steps using these solutions Sodium
hydroxide (SDS), TE buffer containing RNAse, potassium acetate, and ethyl alcohol wash
buffer. Restriction digestion of the isolated plasmid occurred in the step where it was put into the
tube containing the HindIII restriction enzyme. To analyze the digested plasmid agarose gel
electrophoresis was done to determine whether digestion took place. From the results obtained
from gel electrophoresis, we can identify the undigested and digested plasmid. The alkaline lysis
method is very important in the field of biotechnology because it’s a faster and cleaner method of
isolating plasmid DNA from cells. This is because of the plasmid DNA characteristic to anneal
after denaturation (Antipov et al., 2016)
RESULTS.
Legends of the well Starting from the left of the gel image:
What is in the sample for each well?
1) 5 uL H2O, 5 uL DNA, 2 uL 6x LD (loading dye)
2) 3.2 uL H2O, 0.2 uL RNase, 5 uL DNA, 1 uL 10 x Buffer, enzyme or HindIII 0.5
uL
3) 3.5 uL H2O, 5 uL DNA, 1 uL 10 x Buffer, enzyme or HindIII 0.5 uL
4) N

PLASMID ISOLATION. 3
5) N
6) 5 uL DNA, 5 uL H2O, 2 uL 6x LD
7) 5 uL DNA, 5 uL H2O, 2 ul 6x LD
8) 5 uL DNA, 5 uL H2O, 2 ul 6x LD
Student 1: 33.9 ug/ml measured at 260 absorbance
Student 2: 49.6 ug/ml at 260 absorbance
From the above data and calculations in the appendix each student added in each well
Student 1: 0.1695 ug of DNA
Student 2: 0.0248 ug of DNA (see appendix)
5) N
6) 5 uL DNA, 5 uL H2O, 2 uL 6x LD
7) 5 uL DNA, 5 uL H2O, 2 ul 6x LD
8) 5 uL DNA, 5 uL H2O, 2 ul 6x LD
Student 1: 33.9 ug/ml measured at 260 absorbance
Student 2: 49.6 ug/ml at 260 absorbance
From the above data and calculations in the appendix each student added in each well
Student 1: 0.1695 ug of DNA
Student 2: 0.0248 ug of DNA (see appendix)
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
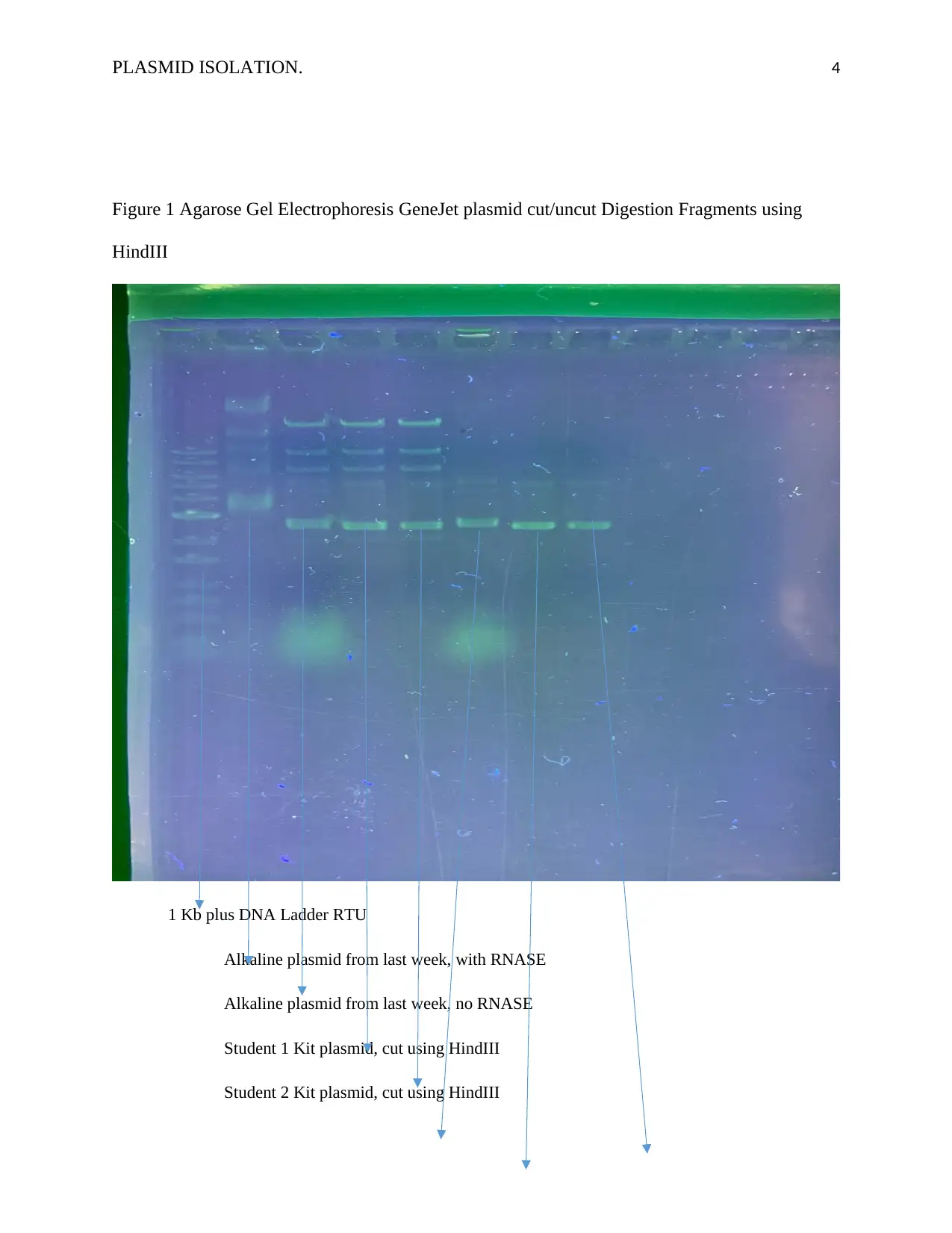
PLASMID ISOLATION. 4
Figure 1 Agarose Gel Electrophoresis GeneJet plasmid cut/uncut Digestion Fragments using
HindIII
1 Kb plus DNA Ladder RTU
Alkaline plasmid from last week, with RNASE
Alkaline plasmid from last week, no RNASE
Student 1 Kit plasmid, cut using HindIII
Student 2 Kit plasmid, cut using HindIII
Figure 1 Agarose Gel Electrophoresis GeneJet plasmid cut/uncut Digestion Fragments using
HindIII
1 Kb plus DNA Ladder RTU
Alkaline plasmid from last week, with RNASE
Alkaline plasmid from last week, no RNASE
Student 1 Kit plasmid, cut using HindIII
Student 2 Kit plasmid, cut using HindIII
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

PLASMID ISOLATION. 5
Alkaline plasmid, no cut, no RNASE (0.1695ug) (0.0248 ug)
Student 1 Kit plasmid, no cut Student 2 Kit plasmid, no cut
THEORY
Question 8. How genetic ladders are created.
A DNA ladder is a solution made up of fragments of different molecular weights, which are
double-stranded. They are used in determining the fragment size of other unknown digested
fragments. Three methods are used in manufacturing a DNA ladder which includes: PCR
amplification method, ligation method, and restriction digestion. In ligation method (a traditional
method and common) phosphodiester bonds are used to join together covalently different
fragments of 100bp to 1000bp (Mostaan et al., 2015)
Question 9.
Manufactures name, Expiry date, generation type
Need of using verified Manufactures of DNA isolation kits.
Ensure the kit's components are of high quality and not counterfeit. This ensures the kits being
used are safe and efficient to give the expected results.
Question 10.
Chaotropic agents serve as proteinase enzymes. They are used in DNA isolation to denature
proteins. Therefore help in getting rid of cell proteins in the DNA isolation and purification
process. In this mini prep kit used for this experiment, potassium acetate and ethyl alcohol wash
buffer is used as a chaotropic agent (Ramakrishnan et al., 2016)
Question 11
A) In a scenario where the initial pellet is not resuspended first before adding solutions 1, 2, 3.
After plasmid isolation is successful one may get a lower quality plasmid as cells won’t be
Alkaline plasmid, no cut, no RNASE (0.1695ug) (0.0248 ug)
Student 1 Kit plasmid, no cut Student 2 Kit plasmid, no cut
THEORY
Question 8. How genetic ladders are created.
A DNA ladder is a solution made up of fragments of different molecular weights, which are
double-stranded. They are used in determining the fragment size of other unknown digested
fragments. Three methods are used in manufacturing a DNA ladder which includes: PCR
amplification method, ligation method, and restriction digestion. In ligation method (a traditional
method and common) phosphodiester bonds are used to join together covalently different
fragments of 100bp to 1000bp (Mostaan et al., 2015)
Question 9.
Manufactures name, Expiry date, generation type
Need of using verified Manufactures of DNA isolation kits.
Ensure the kit's components are of high quality and not counterfeit. This ensures the kits being
used are safe and efficient to give the expected results.
Question 10.
Chaotropic agents serve as proteinase enzymes. They are used in DNA isolation to denature
proteins. Therefore help in getting rid of cell proteins in the DNA isolation and purification
process. In this mini prep kit used for this experiment, potassium acetate and ethyl alcohol wash
buffer is used as a chaotropic agent (Ramakrishnan et al., 2016)
Question 11
A) In a scenario where the initial pellet is not resuspended first before adding solutions 1, 2, 3.
After plasmid isolation is successful one may get a lower quality plasmid as cells won’t be

PLASMID ISOLATION. 6
subjected to the active components of the solutions such as EDTA or RNAse. The role of the
EDTA is to inhibit DNase which denatures plasmid DNA. Hence failure to do resuspension of
the initial pellet will overall affect the quantity and quality of plasmid DNA isolated
B) The potassium acetate role is to renature plasmid DNA while isolating the genomic DNA
together with protein particles, allowing only plasmid DNA to be found within the supernatant.
Therefore this means in case solution 3 does not contain potassium acetate, genomic DNA and
proteins won’t be precipitated. This will make the solution turbid and white and make it very
difficult to analyze bands after gel electrophoresis
C) 70% ethanol is used to remove excess salt impurities from the DNA. Using 95% of ethanol
will precipitate DNA together with impurities instead.
Question 12. Causes of “Ghost bands”.
The appearance of extra bands may due to incomplete digestion as a result of the enzyme not
working as expected, inefficient, or time taken for digestion was shorter than the required timing.
Where bands appear in sizes were not expected may due that the DNA is stuck at different
degrees of supercoiling or digestion. Faint bands may be due to less of the DNA sample
concentration used in setting up the gel electrophoresis. In the case of our experiment, the band
appeared as expected (Dunn, 2014)
Question 14
For Type IIP restriction enzymes, the domain of the amino acid that is involved in catalyzing
cleavage and recognizing the DNA is placed in a single domain which cannot be subdivided. To
the contrast Type IIS restriction enzymes, these domains are placed into two separate domains
joined by polypeptide bond. This makes Type IIS larger than Type IIP enzymes, approximately
400-600 amino acid sequence in length.
subjected to the active components of the solutions such as EDTA or RNAse. The role of the
EDTA is to inhibit DNase which denatures plasmid DNA. Hence failure to do resuspension of
the initial pellet will overall affect the quantity and quality of plasmid DNA isolated
B) The potassium acetate role is to renature plasmid DNA while isolating the genomic DNA
together with protein particles, allowing only plasmid DNA to be found within the supernatant.
Therefore this means in case solution 3 does not contain potassium acetate, genomic DNA and
proteins won’t be precipitated. This will make the solution turbid and white and make it very
difficult to analyze bands after gel electrophoresis
C) 70% ethanol is used to remove excess salt impurities from the DNA. Using 95% of ethanol
will precipitate DNA together with impurities instead.
Question 12. Causes of “Ghost bands”.
The appearance of extra bands may due to incomplete digestion as a result of the enzyme not
working as expected, inefficient, or time taken for digestion was shorter than the required timing.
Where bands appear in sizes were not expected may due that the DNA is stuck at different
degrees of supercoiling or digestion. Faint bands may be due to less of the DNA sample
concentration used in setting up the gel electrophoresis. In the case of our experiment, the band
appeared as expected (Dunn, 2014)
Question 14
For Type IIP restriction enzymes, the domain of the amino acid that is involved in catalyzing
cleavage and recognizing the DNA is placed in a single domain which cannot be subdivided. To
the contrast Type IIS restriction enzymes, these domains are placed into two separate domains
joined by polypeptide bond. This makes Type IIS larger than Type IIP enzymes, approximately
400-600 amino acid sequence in length.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

PLASMID ISOLATION. 7
Type: Acul, Bael
Question 15
Exonucleases.
Exonucleases target and digest nucleic acids by cleaving nucleotides one at a time from the Exo
end of a nucleic acid polypeptide chain. They initiate a hydrolytic reaction that breaks bonds at
either the 3’/ 5’ end. Give rise to single monomers after hydrolysis (Li et al., 2014).
Type relevant to the experiment: Exonuclease 1
Endonucleases.
Endonucleases target and cleave nucleic acids by cleaving phosphodiester bonds within the
polynucleotide chain, endo or middle. They don’t require the 3’ to 5’ direction or vice versa.
After hydrolysis, they give rise to oligonucleotide chains, no single monomers (Dunn, 2014)
Type relevant to the experiment: EcoRI
These nucleases are important in the cleaving of DNA into fragments in DNA recombinant DNA
technology. A buffer in the experiment is added to inhibit the action of these enzymes during
DNA isolation to preserve the integrity of our DNA.
Question 16. Fokl restriction enzyme.
Found in flavobacterium okeankoites naturally and isolated for use in molecular laboratories. It’s
a Type IIS restriction endonuclease. It recognizes the 5′-GGATG-3′ sequence and makes 2
cleaves nine nucleotides away starting from the 3’ end of its recognition sequence from the top
side and thirteen nucleotides away from the 5’ end of the bottom strand sequence forming sticky
ends made up of four nucleotide long overhang (Sinaga et al., 2018)
Type: Acul, Bael
Question 15
Exonucleases.
Exonucleases target and digest nucleic acids by cleaving nucleotides one at a time from the Exo
end of a nucleic acid polypeptide chain. They initiate a hydrolytic reaction that breaks bonds at
either the 3’/ 5’ end. Give rise to single monomers after hydrolysis (Li et al., 2014).
Type relevant to the experiment: Exonuclease 1
Endonucleases.
Endonucleases target and cleave nucleic acids by cleaving phosphodiester bonds within the
polynucleotide chain, endo or middle. They don’t require the 3’ to 5’ direction or vice versa.
After hydrolysis, they give rise to oligonucleotide chains, no single monomers (Dunn, 2014)
Type relevant to the experiment: EcoRI
These nucleases are important in the cleaving of DNA into fragments in DNA recombinant DNA
technology. A buffer in the experiment is added to inhibit the action of these enzymes during
DNA isolation to preserve the integrity of our DNA.
Question 16. Fokl restriction enzyme.
Found in flavobacterium okeankoites naturally and isolated for use in molecular laboratories. It’s
a Type IIS restriction endonuclease. It recognizes the 5′-GGATG-3′ sequence and makes 2
cleaves nine nucleotides away starting from the 3’ end of its recognition sequence from the top
side and thirteen nucleotides away from the 5’ end of the bottom strand sequence forming sticky
ends made up of four nucleotide long overhang (Sinaga et al., 2018)
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

PLASMID ISOLATION. 8

PLASMID ISOLATION. 9
References
Antipov, D., Hartwick, N., Shen, M., Raiko, M., Lapidus, A., & Pevzner, P. A. (2016).
plasmidSPAdes: assembling plasmids from whole genome sequencing data.
Bioinformatics, btw493.
Dehé, P., & Gaillard, P. L. (2017). Control of structure-specific endonucleases to maintain
genome stability. Nature Reviews Molecular Cell Biology, 18(5), 315-330.
Dunn, M. J. (2014). Gel Electrophoresis of Proteins. Elsevier.
Li, S., Chang, H. H., Niewolik, D., Hedrick, M. P., Pinkerton, A. B., Hassig, C. A.,
Schwarz, K., & Lieber, M. R. (2014). Evidence That the DNA Endonuclease
ARTEMIS also Has Intrinsic 5′-Exonuclease Activity. Journal of Biological
Chemistry, 289(11), 7825-7834.
Mostaan, S., Ajorloo, M., Khanahmad, H., Cohan, R., Tehrani, Z., Rezaei, M., Fazeli, F.,
Behdani, M., Zare, S., Karimi, Z., Mozhgani, S., & Moukhah, R. (2015). A novel
combined method for cost-benefit production of DNA ladders. Advanced Biomedical
Research, 4(1), 15.
Ramakrishnan, S., Krainer, G., Grundmeier, G., Schlierf, M., & Keller, A. (2016). Structural
stability of DNA origami nanostructures in the presence of chaotropic agents.
Nanoscale, 8(19), 10398-10405.
Sinaga, B. Y., Amir, Z., & Siagian, P. (2018). The role of FokI polymorphism of vitamin D
receptor gene and vitamin D level in multidrug-resistant tuberculosis occurrence in
Medan city, Indonesia. Archives of Medical Science - Civilization Diseases, 3(1),
153-157.
References
Antipov, D., Hartwick, N., Shen, M., Raiko, M., Lapidus, A., & Pevzner, P. A. (2016).
plasmidSPAdes: assembling plasmids from whole genome sequencing data.
Bioinformatics, btw493.
Dehé, P., & Gaillard, P. L. (2017). Control of structure-specific endonucleases to maintain
genome stability. Nature Reviews Molecular Cell Biology, 18(5), 315-330.
Dunn, M. J. (2014). Gel Electrophoresis of Proteins. Elsevier.
Li, S., Chang, H. H., Niewolik, D., Hedrick, M. P., Pinkerton, A. B., Hassig, C. A.,
Schwarz, K., & Lieber, M. R. (2014). Evidence That the DNA Endonuclease
ARTEMIS also Has Intrinsic 5′-Exonuclease Activity. Journal of Biological
Chemistry, 289(11), 7825-7834.
Mostaan, S., Ajorloo, M., Khanahmad, H., Cohan, R., Tehrani, Z., Rezaei, M., Fazeli, F.,
Behdani, M., Zare, S., Karimi, Z., Mozhgani, S., & Moukhah, R. (2015). A novel
combined method for cost-benefit production of DNA ladders. Advanced Biomedical
Research, 4(1), 15.
Ramakrishnan, S., Krainer, G., Grundmeier, G., Schlierf, M., & Keller, A. (2016). Structural
stability of DNA origami nanostructures in the presence of chaotropic agents.
Nanoscale, 8(19), 10398-10405.
Sinaga, B. Y., Amir, Z., & Siagian, P. (2018). The role of FokI polymorphism of vitamin D
receptor gene and vitamin D level in multidrug-resistant tuberculosis occurrence in
Medan city, Indonesia. Archives of Medical Science - Civilization Diseases, 3(1),
153-157.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

PLASMID ISOLATION. 10
Appendix.
Calculation of ug of DNA sample used.
From the concentration obtained:
33.9ug 1000ul
5ul (student 1)
= 0.1695 ug
N//B Similar method is used for student 2
Appendix.
Calculation of ug of DNA sample used.
From the concentration obtained:
33.9ug 1000ul
5ul (student 1)
= 0.1695 ug
N//B Similar method is used for student 2
1 out of 10
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.

