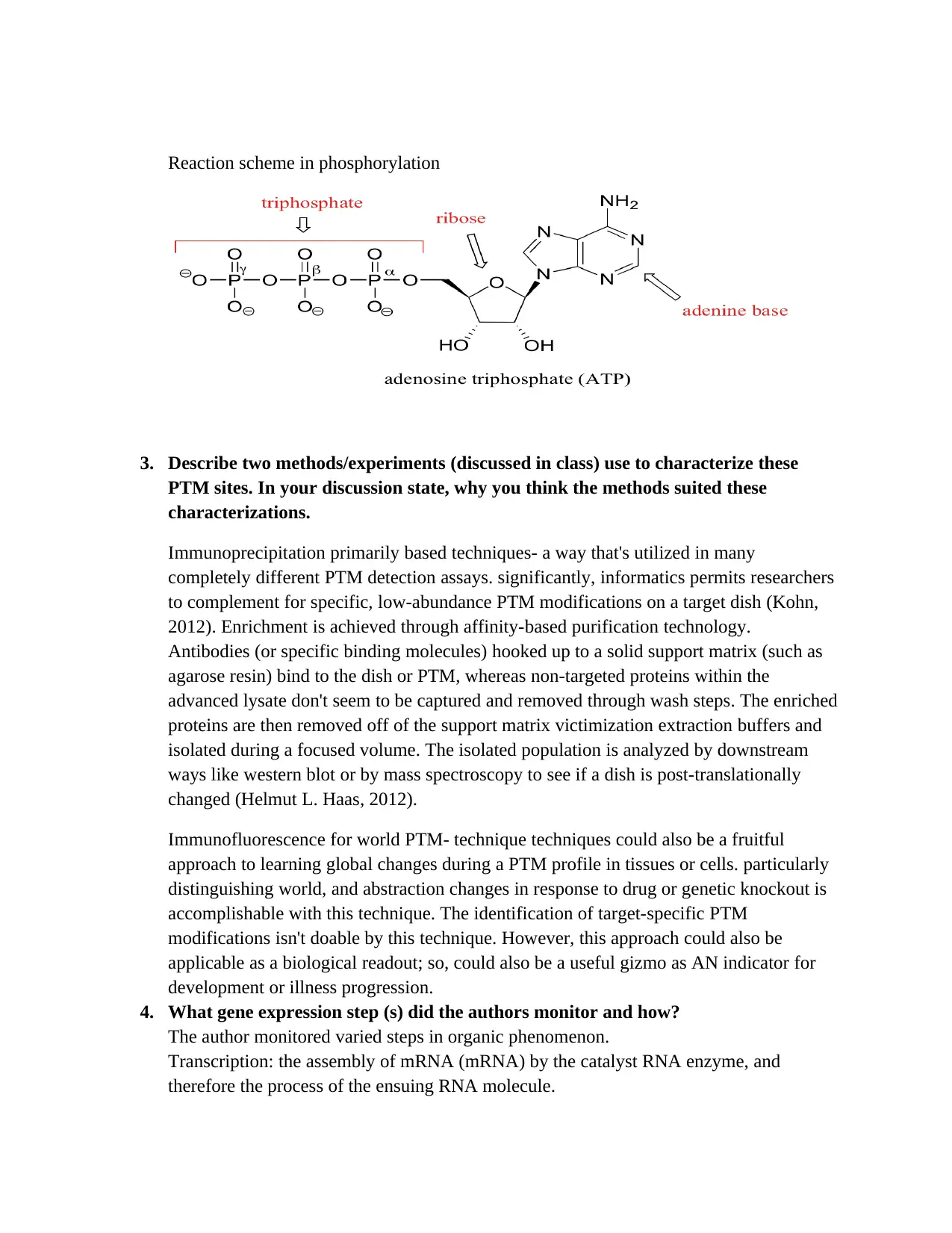
Biochemistry Assignment: PTMs, Gene Expression, Metabolism, and Cancer
VerifiedAdded on 2022/12/28
|4
|874
|75
Homework Assignment
AI Summary
This biochemistry assignment delves into the intricate world of post-translational modifications (PTMs) and their profound impact on gene expression, metabolism, and cancer development. The assignment begins by identifying writer, eraser, and reader proteins, along with their respective enzymes and substrates. It then explores various PTMs, including phosphorylation, protein glycosylation, and ubiquitination, detailing their sites of modification and providing a reaction scheme for one such PTM. Furthermore, the assignment examines two experimental methods, namely immunoprecipitation and immunofluorescence, used to characterize these PTM sites, and explains the rationale behind their suitability. The authors monitored transcription and translation. In summary, the authors' findings suggest that PTMs of biological enzymes contribute to the increase in cancerous cells and metabolism by impacting molecular and genetic processes. The authors link PTMs to metabolism and cancer by showing that these modifications affect metabolic enzymes and the binding of genetic compounds, ultimately influencing cancer metabolism. The findings support the histone code by demonstrating that the protein code does not function as an independent signal to specify the interaction with downstream regulatory proteins, but rather provides further interactions to stabilize the achievement of corepressor complexes.
1 out of 4









![[object Object]](/_next/static/media/star-bottom.7253800d.svg)