Genomic DNA Library Creation: Recombinant DNA Technology
VerifiedAdded on 2020/04/13
|16
|2455
|49
Report
AI Summary
This report details the creation of a genomic DNA library using Bacillus subtilis. The study involved isolating genomic DNA, digesting it with restriction enzymes, and ligating the fragments into the pUC18 vector. The recombinant plasmids were then transformed into E. coli DH5α cells. Blue-white screening was used to identify colonies with successful insertion of the DNA fragments. The results showed a higher ratio of white colonies, indicating successful insertion of the B. subtilis DNA. Restriction digestion confirmed the presence of the insert. The report also includes a discussion of the results, potential problems encountered, and future experiments. The genomic DNA library created can be used to identify genes expressing commercially valuable protein products, identify genetic alterations, and identify genetic regulators.

Running head: RECOMBINANT DNA TECHNOLOGY
Course Code:
RECOMBINANT DNA TECHNOLOGY
Name of the Student:
Name of the Partner:
Name of TA:
Lab Section:
Report Submission Date:
Course Code:
RECOMBINANT DNA TECHNOLOGY
Name of the Student:
Name of the Partner:
Name of TA:
Lab Section:
Report Submission Date:
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

1RECOMBINANT DNA TECHNOLOGY
Abstract
This study was aimed at creating a genomic DNA library. Such a library helps the researcher to
identify the desired genes. The desired gene can be expressed to obtain the protein product. In
this study the Bacillus subtilis genomic DNA was used to create the genomic DNA library. The
genomic and plasmid DNA pUC18 were isolated and their concentrations were determined. The
genomic and plasmid DNA were then subjected to digestion, followed by ligation and
transformed into the Escherichia coli DH5α cells on media containing X-gal/IPTG. The blue
colonies observed lacked the presence of the insert, while the white colonies had the desired
insert ligated into the pUC18 vector. The recombinant plasmids isolated from the recombinant or
white clones were digested with the same restriction enzymes. Double digestion resulted in the
generation of the desired vector and the insert. Thus, the creation of the genomic DNA library
was successful.
Abstract
This study was aimed at creating a genomic DNA library. Such a library helps the researcher to
identify the desired genes. The desired gene can be expressed to obtain the protein product. In
this study the Bacillus subtilis genomic DNA was used to create the genomic DNA library. The
genomic and plasmid DNA pUC18 were isolated and their concentrations were determined. The
genomic and plasmid DNA were then subjected to digestion, followed by ligation and
transformed into the Escherichia coli DH5α cells on media containing X-gal/IPTG. The blue
colonies observed lacked the presence of the insert, while the white colonies had the desired
insert ligated into the pUC18 vector. The recombinant plasmids isolated from the recombinant or
white clones were digested with the same restriction enzymes. Double digestion resulted in the
generation of the desired vector and the insert. Thus, the creation of the genomic DNA library
was successful.

2RECOMBINANT DNA TECHNOLOGY
Table of Contents
Introduction......................................................................................................................................3
Results..............................................................................................................................................4
Lab 3 results.................................................................................................................................4
Lab 4 results.................................................................................................................................4
Lab 5 results.................................................................................................................................5
Discussion........................................................................................................................................7
Reference List..................................................................................................................................9
Appendix........................................................................................................................................11
Table of Contents
Introduction......................................................................................................................................3
Results..............................................................................................................................................4
Lab 3 results.................................................................................................................................4
Lab 4 results.................................................................................................................................4
Lab 5 results.................................................................................................................................5
Discussion........................................................................................................................................7
Reference List..................................................................................................................................9
Appendix........................................................................................................................................11
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

3RECOMBINANT DNA TECHNOLOGY
Introduction
Bacillus subtilis is a Gram-positive non-pathogenic bacillus involved in the formation of
heat resistant and dormant spores (Logan, Niall and Paul). It is the most characterized
microorganisms among the various Gram-positive bacteria present. It genetic material can easily
be subjected to manipulation and as a result widely used in genetic engineering. It is used
commercially for the production of a variety of enzymes, vitamins, flavoring agents and in the
production of industrial nucleotides (Capozzi et al.).
Genomic DNA libraries carry the entire genomic DNA sequence of the organism. The
genomic DNA library can be used to determine the whole genome sequence of an organism
helps to determine the phenotypes regulated by the genes, determination of mutations present in
the genome and in the production of proteins expressed from the respective genes for
commercial use.
Genetic engineering requires the use of recombinant DNA technology in order to carry
out genetic manipulation of an organism. Genetic engineering can also be used to generate
mutations of genes, whose functions are not known (Gaj, Gersbach, and Barbas). Various types
of products like insulin, industrial enzymes and even the human growth hormone has been
produced by the use of genetically modified organisms (GMOs). Genetically modified crops can
also be produced using this technique (Bawa and Anilakumar). The overall purpose of this report
is “to generate the genomic DNA library of B. subtilis and carrying out the necessary steps to
determine its efficacy”.
Introduction
Bacillus subtilis is a Gram-positive non-pathogenic bacillus involved in the formation of
heat resistant and dormant spores (Logan, Niall and Paul). It is the most characterized
microorganisms among the various Gram-positive bacteria present. It genetic material can easily
be subjected to manipulation and as a result widely used in genetic engineering. It is used
commercially for the production of a variety of enzymes, vitamins, flavoring agents and in the
production of industrial nucleotides (Capozzi et al.).
Genomic DNA libraries carry the entire genomic DNA sequence of the organism. The
genomic DNA library can be used to determine the whole genome sequence of an organism
helps to determine the phenotypes regulated by the genes, determination of mutations present in
the genome and in the production of proteins expressed from the respective genes for
commercial use.
Genetic engineering requires the use of recombinant DNA technology in order to carry
out genetic manipulation of an organism. Genetic engineering can also be used to generate
mutations of genes, whose functions are not known (Gaj, Gersbach, and Barbas). Various types
of products like insulin, industrial enzymes and even the human growth hormone has been
produced by the use of genetically modified organisms (GMOs). Genetically modified crops can
also be produced using this technique (Bawa and Anilakumar). The overall purpose of this report
is “to generate the genomic DNA library of B. subtilis and carrying out the necessary steps to
determine its efficacy”.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

4RECOMBINANT DNA TECHNOLOGY
Results
Lab 3 results
The concentration of the B. subtilis genomic DNA isolated in Lab 2 was 44ng/μl. EcoRI
and HindIII restriction enzymes were used to digest the genomic DNA. The digested fragments
were then ligated into the EcoRI and HindIII digested empty vector pUC18 to obtain
recombinant plasmids carrying the various genetic regions of the B. subtilis genome. The
recombinant plasmids thus obtained were introduced into the Escherichia coli DH5α cells.
Restriction digestion of the genomic and plasmid DNA was carried out at 37ºC for 1 hour, then
incubation at 80 ºC for 10 minutes was carried out to inactivate the restriction enzymes. The
digestion products were at first heat inactivated at 45 ºC for 5 minutes to remove the reannealed
digested products. This was followed by the actual ligation step at 18 ºC for 30 minutes. The
ligation products were incubated at 65 ºC for 10 minutes to inactivate the ligase. The ligated
products were transformed into E. coli DH5α cells plated on X-gal, IPTG and Ampicillin
containing LB agar plates.
Lab 4 results
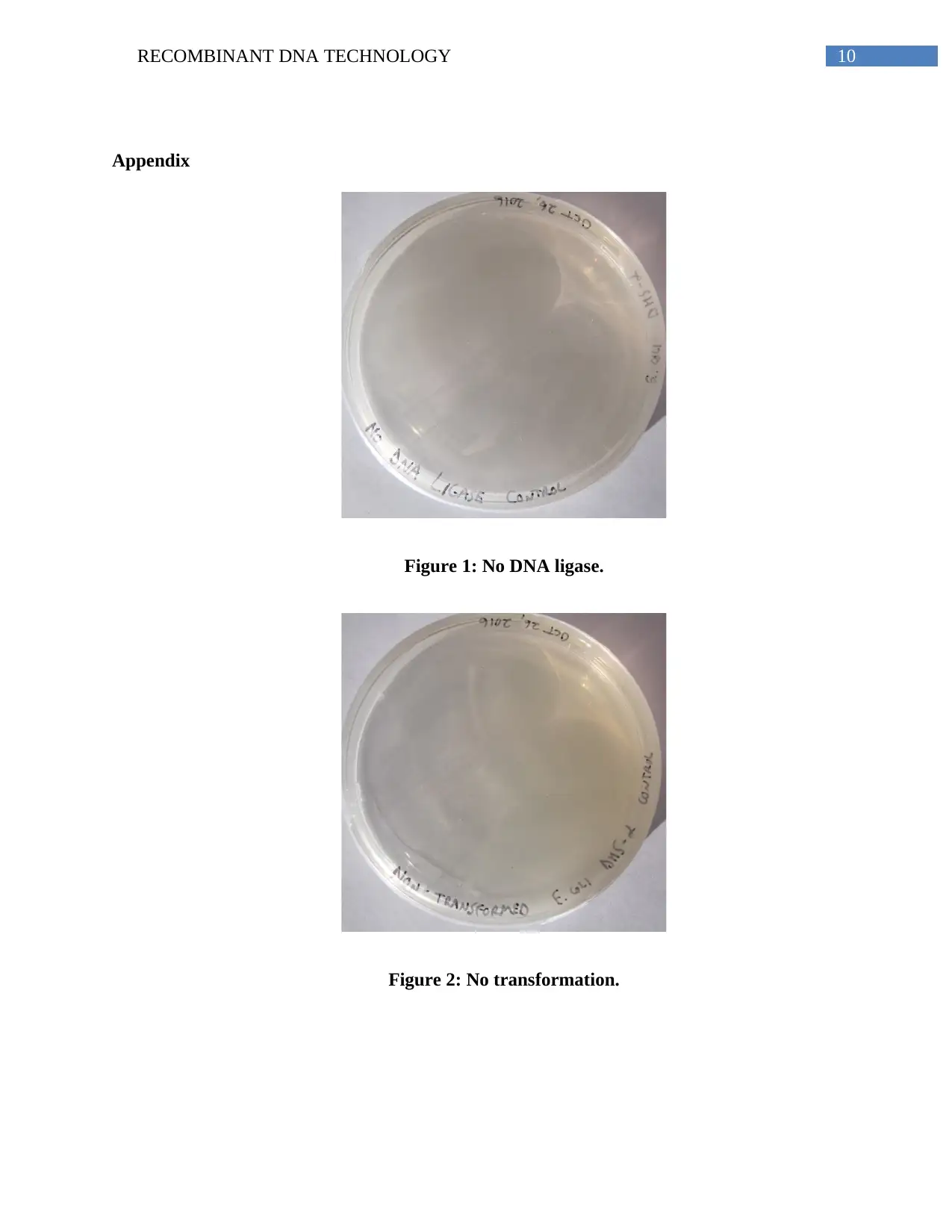
The ratio of the blue to white colonies were 3:8. The control plates used were the
digested and re-ligated pUC18 plates, which gave rise to blue colonies due to the absence of an
insert. The non-transformed and no T4 DNA ligase plates did not show the presence of colonies,
while an already prepared recombinant plasmid was used as the positive control, which gave rise
to white colonies (Apppendix, Figures 1-4). The recombinant clones obtained in the
experimental plates were then used to isolate the recombinant plasmid. A white recombinant
colony was selected. The concentration of the isolated plasmid DNA was 621.1ng/μl and the
Results
Lab 3 results
The concentration of the B. subtilis genomic DNA isolated in Lab 2 was 44ng/μl. EcoRI
and HindIII restriction enzymes were used to digest the genomic DNA. The digested fragments
were then ligated into the EcoRI and HindIII digested empty vector pUC18 to obtain
recombinant plasmids carrying the various genetic regions of the B. subtilis genome. The
recombinant plasmids thus obtained were introduced into the Escherichia coli DH5α cells.
Restriction digestion of the genomic and plasmid DNA was carried out at 37ºC for 1 hour, then
incubation at 80 ºC for 10 minutes was carried out to inactivate the restriction enzymes. The
digestion products were at first heat inactivated at 45 ºC for 5 minutes to remove the reannealed
digested products. This was followed by the actual ligation step at 18 ºC for 30 minutes. The
ligation products were incubated at 65 ºC for 10 minutes to inactivate the ligase. The ligated
products were transformed into E. coli DH5α cells plated on X-gal, IPTG and Ampicillin
containing LB agar plates.
Lab 4 results
The ratio of the blue to white colonies were 3:8. The control plates used were the
digested and re-ligated pUC18 plates, which gave rise to blue colonies due to the absence of an
insert. The non-transformed and no T4 DNA ligase plates did not show the presence of colonies,
while an already prepared recombinant plasmid was used as the positive control, which gave rise
to white colonies (Apppendix, Figures 1-4). The recombinant clones obtained in the
experimental plates were then used to isolate the recombinant plasmid. A white recombinant
colony was selected. The concentration of the isolated plasmid DNA was 621.1ng/μl and the

5RECOMBINANT DNA TECHNOLOGY
260:280 ratio was 2.12. The isolated recombinant plasmid was single digested with HindIII and
double digested with EcoRI/HindIII. This helped to confirm the presence of the insert.
Lab 5 results
The distances travelled by the bands in the DNA ladder were calculated (Appendix, Table
1, Figure 5) and plotted with respect to the length of the DNA bands in base pairs. The X- axis of
the standard curves generated represents the DNA length in base pairs and the Y- axis represents
the distance travelled in mm. A logarithmic trendline was generated in case of both the standard
curves (Appendix, Figures 6 and 7). Calculations using the equations in the standard curves
were done to determine the sizes of the DNA bands in the gel (Appendix, Figure 5). The sizes of
the DNA bands are provided in Table 2 (Appendix).
In gel 1, the pUC 18 plasmid had three bands of sizes 12088, 5115 and 4023bp,
respectively. The double digested pUC18 and the genomic DNA lanes showed one band of sizes
approximately 2697bp and 13359bp, respectively. The control uncut recombinant plasmid had 4
bands of sizes 12088, 5115, 4023 and 2440.6bp. The single digested plasmid had one band of
size 4769bp. The double digested control plasmid produces 2 bands of sizes 3165 and 1353bp,
respectively (Appendix, Figure 5). In gel 3, the genomic DNA had one band of size 11849bp.
The uncut plasmid had 2 bands of sizes 3605 and 2208bp. The single digested plasmid had one
band of size 3605bp. The double digested product had 2 bands of sizes 2697 and 916bp. The
foreign DNA insert was approximately 916bp (Appendix, Figure 5). While the single digested
product was 3605bp approximately, the double digested product adds up to 3613bp. The single
and double digestion of the control recombinant plasmid yielded one and two bands, respectively
(Appendix, Figure 5).
260:280 ratio was 2.12. The isolated recombinant plasmid was single digested with HindIII and
double digested with EcoRI/HindIII. This helped to confirm the presence of the insert.
Lab 5 results
The distances travelled by the bands in the DNA ladder were calculated (Appendix, Table
1, Figure 5) and plotted with respect to the length of the DNA bands in base pairs. The X- axis of
the standard curves generated represents the DNA length in base pairs and the Y- axis represents
the distance travelled in mm. A logarithmic trendline was generated in case of both the standard
curves (Appendix, Figures 6 and 7). Calculations using the equations in the standard curves
were done to determine the sizes of the DNA bands in the gel (Appendix, Figure 5). The sizes of
the DNA bands are provided in Table 2 (Appendix).
In gel 1, the pUC 18 plasmid had three bands of sizes 12088, 5115 and 4023bp,
respectively. The double digested pUC18 and the genomic DNA lanes showed one band of sizes
approximately 2697bp and 13359bp, respectively. The control uncut recombinant plasmid had 4
bands of sizes 12088, 5115, 4023 and 2440.6bp. The single digested plasmid had one band of
size 4769bp. The double digested control plasmid produces 2 bands of sizes 3165 and 1353bp,
respectively (Appendix, Figure 5). In gel 3, the genomic DNA had one band of size 11849bp.
The uncut plasmid had 2 bands of sizes 3605 and 2208bp. The single digested plasmid had one
band of size 3605bp. The double digested product had 2 bands of sizes 2697 and 916bp. The
foreign DNA insert was approximately 916bp (Appendix, Figure 5). While the single digested
product was 3605bp approximately, the double digested product adds up to 3613bp. The single
and double digestion of the control recombinant plasmid yielded one and two bands, respectively
(Appendix, Figure 5).
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

6RECOMBINANT DNA TECHNOLOGY
Discussion
This report describes the preparation of a genomic DNA library. Digestion and ligation of
the genomic DNA inserts into the vector pUC18 yielded the recombinant clones. The number of
white colonies were more than the number of blue colonies. The recombinant plasmid containing
colonies were white because the lacZ gene present in the multiple cloning site (MCS) of the
vector gets disrupted due to the addition of the insert (Davis). The blue colonies carried the
empty vectors, which were obtained due to their undergoing only single digestion. As a result,
they were able to break down the X-gal substrate giving rise to the blue color. This is because the
lacZ gene remained intact producing functional beta galactosidase. Moreover, the double
digestion of the recombinant plasmid yielded DNA bands of sizes 2697 and 916bp, which are the
vector and insert bands, respectively. Additionally, the single digested product gave a single
band of size 3605bp.
The problems that were encountered were the absence of colonies. Absence of colonies
can be due to improper plasmid and genomic DNA purification, which can hamper the digestion
step (Surzycki). Star activity can result in non-specific digestion of the genomic and plasmid
DNAs (Pingoud, Wilson and Wende). Moreover, restriction enzyme inactivation is crucial
otherwise it can interfere with the ligation steps. Lastly, proper generation of competent cells are
necessary for successful transformation (Tu et al.). Other interesting results that were obtained is
the presence of white colonies in the pUC 18 plates and blue colonies in the experimental plate.
The blue colonies in the experimental plate indicates the inappropriate digestion of any one of
the enzymes, thereby resulting in re-ligation of the vector. The control plate containing pUC18 is
expected to produce blue colonies, however, mutations in the lacZ gene can case the appearance
of white colonies.
Discussion
This report describes the preparation of a genomic DNA library. Digestion and ligation of
the genomic DNA inserts into the vector pUC18 yielded the recombinant clones. The number of
white colonies were more than the number of blue colonies. The recombinant plasmid containing
colonies were white because the lacZ gene present in the multiple cloning site (MCS) of the
vector gets disrupted due to the addition of the insert (Davis). The blue colonies carried the
empty vectors, which were obtained due to their undergoing only single digestion. As a result,
they were able to break down the X-gal substrate giving rise to the blue color. This is because the
lacZ gene remained intact producing functional beta galactosidase. Moreover, the double
digestion of the recombinant plasmid yielded DNA bands of sizes 2697 and 916bp, which are the
vector and insert bands, respectively. Additionally, the single digested product gave a single
band of size 3605bp.
The problems that were encountered were the absence of colonies. Absence of colonies
can be due to improper plasmid and genomic DNA purification, which can hamper the digestion
step (Surzycki). Star activity can result in non-specific digestion of the genomic and plasmid
DNAs (Pingoud, Wilson and Wende). Moreover, restriction enzyme inactivation is crucial
otherwise it can interfere with the ligation steps. Lastly, proper generation of competent cells are
necessary for successful transformation (Tu et al.). Other interesting results that were obtained is
the presence of white colonies in the pUC 18 plates and blue colonies in the experimental plate.
The blue colonies in the experimental plate indicates the inappropriate digestion of any one of
the enzymes, thereby resulting in re-ligation of the vector. The control plate containing pUC18 is
expected to produce blue colonies, however, mutations in the lacZ gene can case the appearance
of white colonies.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

7RECOMBINANT DNA TECHNOLOGY
Additional experiments includes polymerase chain and cycle sequencing to confirm the
presence of the insert in the desired vector (Hoseini and Sauer).
The genomic DNA library can be used identify genes that express commercially valuable
protein products. Genomic DNA libraries can also be used to identify genetic alterations of an
organism and also identify the genetic regulators that modulate an organism’s genetic circuitry.
The purpose is to overexpress the desired gene and produce large quantities of proteins,
which will be subjected to further purification.
The blue white screening is based on the theory of α-complementation. lacZ encodes the
enzyme β-galactosidase, which is a tetramer having 2 α and 2 ω fragments. E. coli cells that lack
the α fragment, produce non-functional β-galactosidase. However, the α fragment can be
introduced by the introduction of a plasmid expressing the α fragment in trans. The lacZ gene
present in the MCS gets disrupted by the addition of an insert (Blau and Wehrman). IPTG and X-
gal is added to the LB media. IPTG acts as the gratuitous inducer and X-gal functions as the
chromogenic substrate. Non-functional LacZ cannot degrade the substrate and produce white
coloration, while functional LacZ degrades the substrate to produce blue coloration. The
recombinant plasmids are identified by the white coloration of the colonies (Chaudhuri). DH5α
cells are ΔM15 strains, where 11-41 amino acid residues (α fragment) from the N-terminal of
LacZ is deleted and subsequently the residual ω fragment is inactive. Thus, the DH5α strain is
suitable for such a screening as blue colonies will appear only when a plasmid expressing the α
fragment is introduced (Aguilera and Aguilera-Gómez).
Additional experiments includes polymerase chain and cycle sequencing to confirm the
presence of the insert in the desired vector (Hoseini and Sauer).
The genomic DNA library can be used identify genes that express commercially valuable
protein products. Genomic DNA libraries can also be used to identify genetic alterations of an
organism and also identify the genetic regulators that modulate an organism’s genetic circuitry.
The purpose is to overexpress the desired gene and produce large quantities of proteins,
which will be subjected to further purification.
The blue white screening is based on the theory of α-complementation. lacZ encodes the
enzyme β-galactosidase, which is a tetramer having 2 α and 2 ω fragments. E. coli cells that lack
the α fragment, produce non-functional β-galactosidase. However, the α fragment can be
introduced by the introduction of a plasmid expressing the α fragment in trans. The lacZ gene
present in the MCS gets disrupted by the addition of an insert (Blau and Wehrman). IPTG and X-
gal is added to the LB media. IPTG acts as the gratuitous inducer and X-gal functions as the
chromogenic substrate. Non-functional LacZ cannot degrade the substrate and produce white
coloration, while functional LacZ degrades the substrate to produce blue coloration. The
recombinant plasmids are identified by the white coloration of the colonies (Chaudhuri). DH5α
cells are ΔM15 strains, where 11-41 amino acid residues (α fragment) from the N-terminal of
LacZ is deleted and subsequently the residual ω fragment is inactive. Thus, the DH5α strain is
suitable for such a screening as blue colonies will appear only when a plasmid expressing the α
fragment is introduced (Aguilera and Aguilera-Gómez).

8RECOMBINANT DNA TECHNOLOGY
Reference List
Aguilera, Margarita, and Jesús Manuel Aguilera-Gómez. "chapter seventeen." Applied Molecular
Biotechnology: The Next Generation of Genetic Engineering (2016): 405.
Bawa, A. S., and K. R. Anilakumar. "Genetically modified foods: safety, risks and public
concerns—a review." Journal of food science and technology 50.6 (2013): 1035-1046.
Blau, Helen M., and Thomas S. Wehrman. "Detection of protein translocation by beta-
galactosidase reporter fragment complementation." U.S. Patent No. 8,586,294. 19 Nov. 2013.
Capozzi, Vittorio, et al. "Lactic acid bacteria producing B-group vitamins: a great potential for
functional cereals products." Applied microbiology and biotechnology 96.6 (2012): 1383-1394.
Chaudhuri, Keya. Recombinant DNA Technology. The Energy and Resources Institute (TERI),
2013.
Davis, Leonard. Basic methods in molecular biology. Elsevier, 2012.
Gaj, Thomas, Charles A. Gersbach, and Carlos F. Barbas. "ZFN, TALEN, and CRISPR/Cas-
based methods for genome engineering." Trends in biotechnology 31.7 (2013): 397-405.
Hoseini, Sayed Shahabuddin, and Martin G. Sauer. "Molecular cloning using polymerase chain
reaction, an educational guide for cellular engineering." Journal of biological engineering 9.1
(2015): 2.
Logan, Niall A., and Paul De Vos. "Bacillus." Bergey's Manual of Systematics of Archaea and
Bacteria (2015).
Reference List
Aguilera, Margarita, and Jesús Manuel Aguilera-Gómez. "chapter seventeen." Applied Molecular
Biotechnology: The Next Generation of Genetic Engineering (2016): 405.
Bawa, A. S., and K. R. Anilakumar. "Genetically modified foods: safety, risks and public
concerns—a review." Journal of food science and technology 50.6 (2013): 1035-1046.
Blau, Helen M., and Thomas S. Wehrman. "Detection of protein translocation by beta-
galactosidase reporter fragment complementation." U.S. Patent No. 8,586,294. 19 Nov. 2013.
Capozzi, Vittorio, et al. "Lactic acid bacteria producing B-group vitamins: a great potential for
functional cereals products." Applied microbiology and biotechnology 96.6 (2012): 1383-1394.
Chaudhuri, Keya. Recombinant DNA Technology. The Energy and Resources Institute (TERI),
2013.
Davis, Leonard. Basic methods in molecular biology. Elsevier, 2012.
Gaj, Thomas, Charles A. Gersbach, and Carlos F. Barbas. "ZFN, TALEN, and CRISPR/Cas-
based methods for genome engineering." Trends in biotechnology 31.7 (2013): 397-405.
Hoseini, Sayed Shahabuddin, and Martin G. Sauer. "Molecular cloning using polymerase chain
reaction, an educational guide for cellular engineering." Journal of biological engineering 9.1
(2015): 2.
Logan, Niall A., and Paul De Vos. "Bacillus." Bergey's Manual of Systematics of Archaea and
Bacteria (2015).
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

9RECOMBINANT DNA TECHNOLOGY
Pingoud, Alfred, Geoffrey G. Wilson, and Wolfgang Wende. "Type II restriction endonucleases
—a historical perspective and more." Nucleic acids research 42.12 (2014): 7489-7527.
Surzycki, Stefan. Basic techniques in molecular biology. Springer Science & Business Media,
2012.
Tu, Qiang, et al. "Room temperature electrocompetent bacterial cells improve DNA
transformation and recombineering efficiency." Scientific reports 6 (2016).
Pingoud, Alfred, Geoffrey G. Wilson, and Wolfgang Wende. "Type II restriction endonucleases
—a historical perspective and more." Nucleic acids research 42.12 (2014): 7489-7527.
Surzycki, Stefan. Basic techniques in molecular biology. Springer Science & Business Media,
2012.
Tu, Qiang, et al. "Room temperature electrocompetent bacterial cells improve DNA
transformation and recombineering efficiency." Scientific reports 6 (2016).
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
10RECOMBINANT DNA TECHNOLOGY
Appendix
Figure 1: No DNA ligase.
Figure 2: No transformation.
Appendix
Figure 1: No DNA ligase.
Figure 2: No transformation.
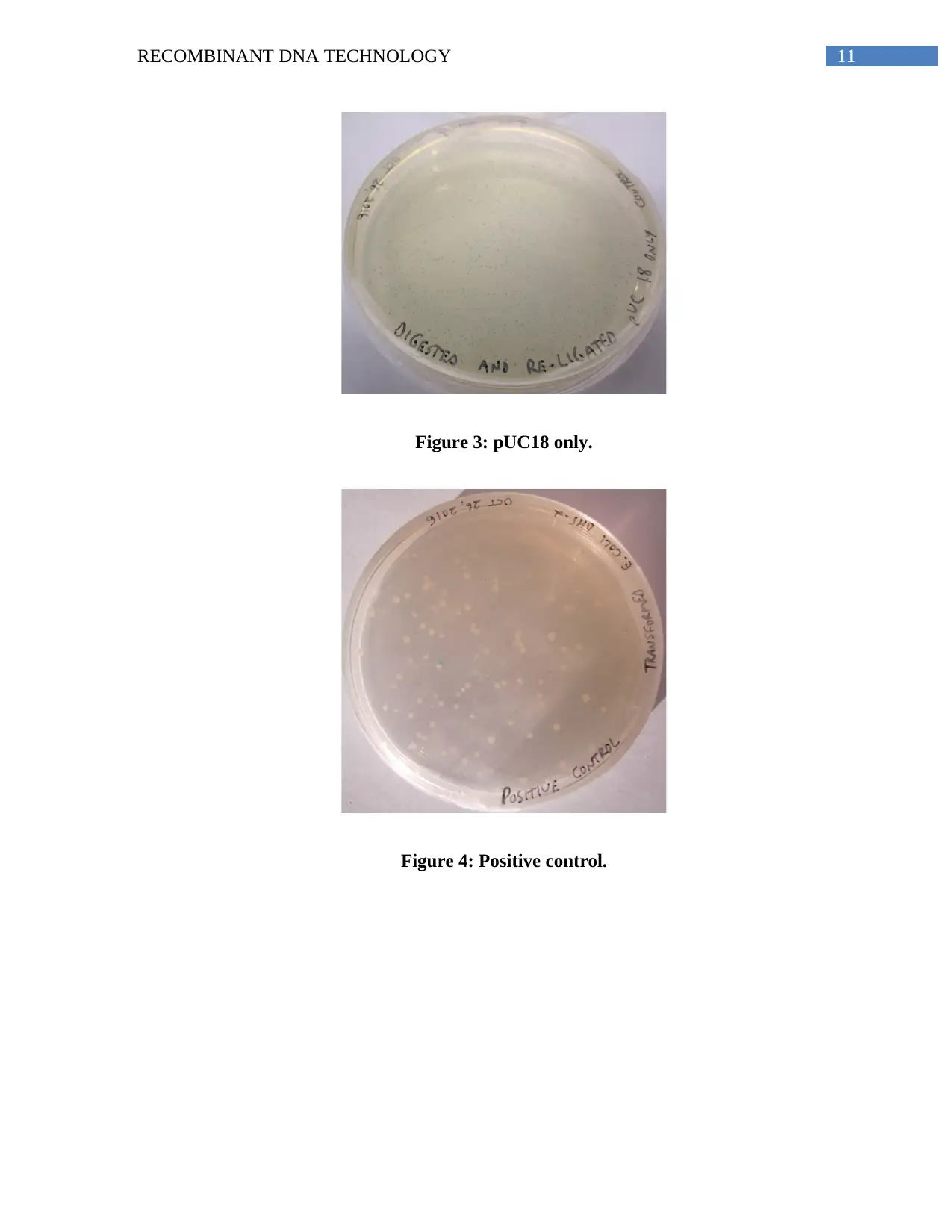
11RECOMBINANT DNA TECHNOLOGY
Figure 3: pUC18 only.
Figure 4: Positive control.
Figure 3: pUC18 only.
Figure 4: Positive control.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 16
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.



