SilE Protein: Characteristics and Role in Bacterial Silver Resistance
VerifiedAdded on 2022/11/16
|21
|5514
|243
Report
AI Summary
This report details a research project investigating the SilE protein, focusing on its characteristics and role in bacterial silver resistance. The study begins with the overexpression and purification of the SilE protein in an E. coli system, followed by structural biochemistry analysis using two-dimensional NMR techniques. The research further explores the silver-induced folding of SilE, employing Nuclear Overhauser Effect or X-ray crystallography to understand the protein's mechanism of action. The project aims to provide insights into the silver resistance mechanism, which could potentially lead to the development of strategies to combat the spread of antibiotic resistance. The results of the experiment are presented which include the purification process and the confirmation of the presence of SilE protein. The report also provides a detailed literature review on SilE and its mechanism of action.

Running Head: SilE- CHARACTERISTICS AND ROLE IN BACTERIAL SILVER
RESISTANCE
SilE: Its characteristics and role in bacterial silver resistance
Name of Student:
Name of University:
Author Note:
RESISTANCE
SilE: Its characteristics and role in bacterial silver resistance
Name of Student:
Name of University:
Author Note:
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

1SilE: Its characteristics and role in bacterial silver resistance
Abstract
The multidrug resistance imparted by bacteria against antibiotics is increasing at a rapid rate
globally that causes several comorbid infections. Silver is also an excellent antibacterial agent
which can be used to treat burns and wounds. But there exists a raising concern about the
resistance imparted to the metal ion. There are several bacterial isolates that are resistant to
silver due to presence of an operon called the Sil operon which consists of an efflux pump
and a protein that binds to silver ions and imparts silver resistance to the system known as
SilE. The objective of the research is to understand the characteristic of the target protein SilE
that involves the silver induced folding of the protein.
The project has three subdivision that involves overexpression of the SilE protein in
the E.coli system and subsequent extraction and purification of the target protein. The second
part of the project deals with the structural biochemistry of the protein of choice exploiting
two-dimensional NMR technique. Lastly, this study deals with the biochemistry involved in
the silver induced folding of SilE. The technique involved is Nuclear Overhauser Effect or X
ray crystallography.
This research study would help to understand the mechanism of action of the protein
SilE and appropriate follow up research studies that would help in reduction and eradication
of the silver resistance.
Table of Content
Abstract
The multidrug resistance imparted by bacteria against antibiotics is increasing at a rapid rate
globally that causes several comorbid infections. Silver is also an excellent antibacterial agent
which can be used to treat burns and wounds. But there exists a raising concern about the
resistance imparted to the metal ion. There are several bacterial isolates that are resistant to
silver due to presence of an operon called the Sil operon which consists of an efflux pump
and a protein that binds to silver ions and imparts silver resistance to the system known as
SilE. The objective of the research is to understand the characteristic of the target protein SilE
that involves the silver induced folding of the protein.
The project has three subdivision that involves overexpression of the SilE protein in
the E.coli system and subsequent extraction and purification of the target protein. The second
part of the project deals with the structural biochemistry of the protein of choice exploiting
two-dimensional NMR technique. Lastly, this study deals with the biochemistry involved in
the silver induced folding of SilE. The technique involved is Nuclear Overhauser Effect or X
ray crystallography.
This research study would help to understand the mechanism of action of the protein
SilE and appropriate follow up research studies that would help in reduction and eradication
of the silver resistance.
Table of Content

2SilE: Its characteristics and role in bacterial silver resistance
s
Introduction...........................................................................................................................3
Antibiotic Resistance Crisis..............................................................................................3
Use of silver in the resistance process...............................................................................3
Resistance to silver............................................................................................................4
Literature review on SilE and its mechanism of action.....................................................5
Materials and Methodology...................................................................................................6
Result.....................................................................................................................................7
Overexpression and purification of SilE...........................................................................7
Characterisation of SilE based on the silver induced folding............................................9
Acquiring the structural information of the silver-bound folded SilE............................11
Discussion...........................................................................................................................12
Conclusion...........................................................................................................................15
References...........................................................................................................................16
s
Introduction...........................................................................................................................3
Antibiotic Resistance Crisis..............................................................................................3
Use of silver in the resistance process...............................................................................3
Resistance to silver............................................................................................................4
Literature review on SilE and its mechanism of action.....................................................5
Materials and Methodology...................................................................................................6
Result.....................................................................................................................................7
Overexpression and purification of SilE...........................................................................7
Characterisation of SilE based on the silver induced folding............................................9
Acquiring the structural information of the silver-bound folded SilE............................11
Discussion...........................................................................................................................12
Conclusion...........................................................................................................................15
References...........................................................................................................................16
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

3SilE: Its characteristics and role in bacterial silver resistance
Introduction
Antibiotic Resistance Crisis
The emergence of multidrug resistance to combat pathogenic bacteria jeopardize the
worth of the antibiotics which had brought about revolutionary changes in the world of
medical science. There are several reasons that impact the antibiotic resistance which
includes misuse of antibiotics, usage of incorrectly prescribed antibiotics and extensive use in
agricultural practice, regulatory barriers and unavailability of new antibiotics (Ventola,
2015). Antibiotic resistance leads to certain comorbid infections that pose as a substantial
burden both economically and in terms of health (Aslam et al, 2018). Multidisciplinary
approaches in the health care, environment and agricultural sectors are required to reduce the
rapid pace of antibiotic resistance by exploiting the antimicrobial agents and the mode of
resistance mechanism.
Use of silver in the resistance process
It is known since ancient times that silver has antimicrobial effects and is effective in
the process of wound healing or stomach pain. It is widely used in hospitals to prevent
nosocomial infections. In Japanese pills silver is widely used to reduce and eradicate bad
breath, nausea, hangover, sunstroke and others. Silver ion is considered an efficacious potent
of different classes of antibiotics and can be effectively used as biocides. Silver nanoparticles
is blooming commercially in the field of nanoparticles used for bactericidal, medical and
electrical product. Silver ion acts on a multiple targets which includes macromolecules, free
amino acid like cysteine, small molecules like glutathione (Maillard & Hartemann, 2013).
This is the reason why it becomes difficult to trace the main reason for the death of a bacteria
treated with silver ions (Barras, Aussel and Ezraty, 2018).
Introduction
Antibiotic Resistance Crisis
The emergence of multidrug resistance to combat pathogenic bacteria jeopardize the
worth of the antibiotics which had brought about revolutionary changes in the world of
medical science. There are several reasons that impact the antibiotic resistance which
includes misuse of antibiotics, usage of incorrectly prescribed antibiotics and extensive use in
agricultural practice, regulatory barriers and unavailability of new antibiotics (Ventola,
2015). Antibiotic resistance leads to certain comorbid infections that pose as a substantial
burden both economically and in terms of health (Aslam et al, 2018). Multidisciplinary
approaches in the health care, environment and agricultural sectors are required to reduce the
rapid pace of antibiotic resistance by exploiting the antimicrobial agents and the mode of
resistance mechanism.
Use of silver in the resistance process
It is known since ancient times that silver has antimicrobial effects and is effective in
the process of wound healing or stomach pain. It is widely used in hospitals to prevent
nosocomial infections. In Japanese pills silver is widely used to reduce and eradicate bad
breath, nausea, hangover, sunstroke and others. Silver ion is considered an efficacious potent
of different classes of antibiotics and can be effectively used as biocides. Silver nanoparticles
is blooming commercially in the field of nanoparticles used for bactericidal, medical and
electrical product. Silver ion acts on a multiple targets which includes macromolecules, free
amino acid like cysteine, small molecules like glutathione (Maillard & Hartemann, 2013).
This is the reason why it becomes difficult to trace the main reason for the death of a bacteria
treated with silver ions (Barras, Aussel and Ezraty, 2018).
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

4SilE: Its characteristics and role in bacterial silver resistance
Silver compounds have wide range of utility as effective antimicrobial agent to fight
against bacteria, viruses and other eukaryotic microbes which are found in the hospital as
well as in public health hygiene. Silver cations are bactericidal at lower concentrations and is
effective in treating ulcers, burns and wounds. Silver is also used to coat catheters to prevent
the development of microbial films. Silver is used in face creams, water filtration cartridges
and supermarket products for washing vegetables. Silver nanoparticles (AgNPs) successfully
caused growth inhibition and oxidative stress in both Gram positive and Gram negative
bacteria, Escherichia coli and Staphylococcus aureus respectively (Singh et al., 2015). But it
was observed that bacteria when pre-exposed to sub-lethal dose of AgNPs showed increased
resistance to ampicillin and penicillin-streptomycin (Pen-Strep). Reduced levels of membrane
damage and reduced oxidative was observed in the pre-exposed bacteria. They also exhibited
high intracellular ATP levels in comparison to the untreated cells. An Ames test was
performed which proved that AgNPs are immensely mutagenic in nature, generates abiotic
reactive oxygen species (ROS) that induces stress tolerance in the bacterial species and also
promotes DNA cleavage in vitro (Kaweeteerawat et al, 2017).
Resistance to silver
The resistance to silver was first encountered in a strain of Salmonella typimurium. It
caused an outbreak on a burn ward and was the reason of death of three people that occurred
in the year 1975 (Percival, Bowler & Russell, 2005). Gram negative bacteria like Escherichia
coli 013, CCM 3954 strain and Pseudomonas aeruginosa CCM 3955 has been observed to
develop resistance to AgNPs on exposure to it repeatedly (Sütterlin, 2015). The resistance
involves phenotypic changes only like production of flagellin, which an adhesive flagellum
protein that initiates the nanoparticle aggregation (Panáček et al, 2018). The changes
collectively reduce the colloidal stability of the nanoparticles and causes decreased
antibacterial activity.
Silver compounds have wide range of utility as effective antimicrobial agent to fight
against bacteria, viruses and other eukaryotic microbes which are found in the hospital as
well as in public health hygiene. Silver cations are bactericidal at lower concentrations and is
effective in treating ulcers, burns and wounds. Silver is also used to coat catheters to prevent
the development of microbial films. Silver is used in face creams, water filtration cartridges
and supermarket products for washing vegetables. Silver nanoparticles (AgNPs) successfully
caused growth inhibition and oxidative stress in both Gram positive and Gram negative
bacteria, Escherichia coli and Staphylococcus aureus respectively (Singh et al., 2015). But it
was observed that bacteria when pre-exposed to sub-lethal dose of AgNPs showed increased
resistance to ampicillin and penicillin-streptomycin (Pen-Strep). Reduced levels of membrane
damage and reduced oxidative was observed in the pre-exposed bacteria. They also exhibited
high intracellular ATP levels in comparison to the untreated cells. An Ames test was
performed which proved that AgNPs are immensely mutagenic in nature, generates abiotic
reactive oxygen species (ROS) that induces stress tolerance in the bacterial species and also
promotes DNA cleavage in vitro (Kaweeteerawat et al, 2017).
Resistance to silver
The resistance to silver was first encountered in a strain of Salmonella typimurium. It
caused an outbreak on a burn ward and was the reason of death of three people that occurred
in the year 1975 (Percival, Bowler & Russell, 2005). Gram negative bacteria like Escherichia
coli 013, CCM 3954 strain and Pseudomonas aeruginosa CCM 3955 has been observed to
develop resistance to AgNPs on exposure to it repeatedly (Sütterlin, 2015). The resistance
involves phenotypic changes only like production of flagellin, which an adhesive flagellum
protein that initiates the nanoparticle aggregation (Panáček et al, 2018). The changes
collectively reduce the colloidal stability of the nanoparticles and causes decreased
antibacterial activity.

5SilE: Its characteristics and role in bacterial silver resistance
Literature review on SilE and its mechanism of action
Microorganisms face a challenge with silver toxicity in medical and environmental
settings. But through repeated exposures bacteria gains adaptation in growing in higher silver
ion concentration. These microbes are a potent threat as a pathogen. Even though
nanoparticles intervention is efficacious but there a naturally adapted microbes exploit efflux
pumps to attain resistance to metals. There are 70 enteric isolates observed from different
hospital locations including catheters and other silver exposed sites (Sedlak et al., 2012). The
molecular genetics which majorly involves bacterial plasmids and genes determine the mode
of resistance against silver compounds. The silver resistance was first observed in Salmonella
typhimurium that was isolated from burn wounds in 1975 (Mirolo et al., 2013). The plasmid
pMGH100 is a Salmonella plasmid that involves a transcription unit consisting of nine genes.
A transcriptional regulatory complex SilRS, which consists of two components sensor and
responder, these governs the synthesis of SilE, a periplasmic silver binding protein, two
efflux pumps, a P-type ATPase SilP and triple protein chemiosmotic in nature, SilCBA which
is a silver-hydrogen ion exchange system (Silver, 2003). This mechanism causes silver
externalization. The resistance of silver is encoded by an operon system called sil operon
observed in pM101. It promotes the export of silver via SilP, SilCFBA and the periplasmic
chaperone protein SilE.
Literature review on SilE and its mechanism of action
Microorganisms face a challenge with silver toxicity in medical and environmental
settings. But through repeated exposures bacteria gains adaptation in growing in higher silver
ion concentration. These microbes are a potent threat as a pathogen. Even though
nanoparticles intervention is efficacious but there a naturally adapted microbes exploit efflux
pumps to attain resistance to metals. There are 70 enteric isolates observed from different
hospital locations including catheters and other silver exposed sites (Sedlak et al., 2012). The
molecular genetics which majorly involves bacterial plasmids and genes determine the mode
of resistance against silver compounds. The silver resistance was first observed in Salmonella
typhimurium that was isolated from burn wounds in 1975 (Mirolo et al., 2013). The plasmid
pMGH100 is a Salmonella plasmid that involves a transcription unit consisting of nine genes.
A transcriptional regulatory complex SilRS, which consists of two components sensor and
responder, these governs the synthesis of SilE, a periplasmic silver binding protein, two
efflux pumps, a P-type ATPase SilP and triple protein chemiosmotic in nature, SilCBA which
is a silver-hydrogen ion exchange system (Silver, 2003). This mechanism causes silver
externalization. The resistance of silver is encoded by an operon system called sil operon
observed in pM101. It promotes the export of silver via SilP, SilCFBA and the periplasmic
chaperone protein SilE.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

6SilE: Its characteristics and role in bacterial silver resistance
Fig 1: Mechanism of Silver resistance by SilE (Asiani et al., 2016)
SilE is a protein which plays an important role in silver detoxification. The primary
structure of the SilE protein is made up of 123 amino acids. This also includes a 20 amino
acid sequence in the N terminal that is cleaved after the periplasm is targeted. The protein is
said to bind to the silver ions with the help of histidine residues. There is a shift observed in
the histidine imidazole proton resonances, in the NMR studies when silver binds to the SilE.
The alpha helix folding of the protein upon binding of the silver molecule also causes
chemical shifts. It is considered that SilE is intrinsically disordered (Asiani et al., 2016). It is
intrinsically disordered when it is in its free apo form but when it is in the holo form, it forms
a compact structure upon optimal binding with approximately six silver ions. Studies based
on sequence analyses and site-directed mutagenesis confirm the significance of the histidine
and methionine containing motifs for silver binding (Zhang, Liang, & Hu, 2014). Relative
studies recognised the presence of a nucleation core that causes silver mediated folding of
SilE. It is considered as a molecular sponge that readily absorbs metal ions.
Materials and Methodology
The Escherichia coli cells were treated with the plasmid pMAL-C5α which had the
SilE gene incorporated in it. This protein was overexpressed in the Escherichia coli strain and
was subsequently extracted using an amylose column. The amylose resin has an affinity for
MBP and the SilE protein extracted is also MBP tagged (Duong & Gabelli, 2015). After the
purification, the protein was cleaved by a protease enzyme called factor Xa. To confirm that
the extracted protein was the desired protein SilE, the different fractions of the purification
were run in the agarose gel. The elution fractions which had the protein of interest was
further concentrated and again run in the agarose gel to reconfirm the result and ensure that
the protein is comparatively pure and contain less junk.
Fig 1: Mechanism of Silver resistance by SilE (Asiani et al., 2016)
SilE is a protein which plays an important role in silver detoxification. The primary
structure of the SilE protein is made up of 123 amino acids. This also includes a 20 amino
acid sequence in the N terminal that is cleaved after the periplasm is targeted. The protein is
said to bind to the silver ions with the help of histidine residues. There is a shift observed in
the histidine imidazole proton resonances, in the NMR studies when silver binds to the SilE.
The alpha helix folding of the protein upon binding of the silver molecule also causes
chemical shifts. It is considered that SilE is intrinsically disordered (Asiani et al., 2016). It is
intrinsically disordered when it is in its free apo form but when it is in the holo form, it forms
a compact structure upon optimal binding with approximately six silver ions. Studies based
on sequence analyses and site-directed mutagenesis confirm the significance of the histidine
and methionine containing motifs for silver binding (Zhang, Liang, & Hu, 2014). Relative
studies recognised the presence of a nucleation core that causes silver mediated folding of
SilE. It is considered as a molecular sponge that readily absorbs metal ions.
Materials and Methodology
The Escherichia coli cells were treated with the plasmid pMAL-C5α which had the
SilE gene incorporated in it. This protein was overexpressed in the Escherichia coli strain and
was subsequently extracted using an amylose column. The amylose resin has an affinity for
MBP and the SilE protein extracted is also MBP tagged (Duong & Gabelli, 2015). After the
purification, the protein was cleaved by a protease enzyme called factor Xa. To confirm that
the extracted protein was the desired protein SilE, the different fractions of the purification
were run in the agarose gel. The elution fractions which had the protein of interest was
further concentrated and again run in the agarose gel to reconfirm the result and ensure that
the protein is comparatively pure and contain less junk.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

7SilE: Its characteristics and role in bacterial silver resistance
The protein was then subjected to Heteronuclear Single Quantum Coherence (HSQC)
which is a two dimensional NMR spectroscopy. But it was observed that there was
comparatively more noise and background and alongside it was difficult to decipher whether
the protein was folded or not.
Thus, to study on the silver induced protein folding X ray crystallography and three
dimensional NMR technique, Nuclear overhauser effect was used to determine the clear
structure and consecutively decipher the characteristic of the desired protein SilE.
Result
Overexpression and purification of SilE
The preliminary step of the research involves cloning of SilE in Escherichia coli cells
(E. coli) and subsequent overexpression of the same. The aim of this experiment was to
ensure optimal conditions for the protein expression and purification.
E.coli cells are cultured and the desired protein is expressed in pMAL-C5α plasmid.
The cloned gene for SilE expression was inserted into the plasmid downstream from the gene
malE which codes for Manose Binding Protein (MBP). This caused formation and expression
of the fusion protein. The fusion protein consisted of SilE tagged MBP. This synthesis is
promoted by the Ptac promoter, which is a strong promoter. The expression of the fusion
protein was checked with the help of BL21-Gold and CodonPlus. This was mainly done to
eliminate the codon bias issue that might obstruct the expression of the fusion protein in
E.coli hosts. The BL21-Gold cells boost the expression of protein by supplying extra copies
of target specific tRNA genes which are not common in the E. coli genome. In BL21 Gold
and Codon Plus the expressions of the fusion protein was checked. Cells were cultured in
minimal media and the expression of the protein was checked at varying temperatures. The
fusion protein produced was purified by single step affinity purification with a resin having
The protein was then subjected to Heteronuclear Single Quantum Coherence (HSQC)
which is a two dimensional NMR spectroscopy. But it was observed that there was
comparatively more noise and background and alongside it was difficult to decipher whether
the protein was folded or not.
Thus, to study on the silver induced protein folding X ray crystallography and three
dimensional NMR technique, Nuclear overhauser effect was used to determine the clear
structure and consecutively decipher the characteristic of the desired protein SilE.
Result
Overexpression and purification of SilE
The preliminary step of the research involves cloning of SilE in Escherichia coli cells
(E. coli) and subsequent overexpression of the same. The aim of this experiment was to
ensure optimal conditions for the protein expression and purification.
E.coli cells are cultured and the desired protein is expressed in pMAL-C5α plasmid.
The cloned gene for SilE expression was inserted into the plasmid downstream from the gene
malE which codes for Manose Binding Protein (MBP). This caused formation and expression
of the fusion protein. The fusion protein consisted of SilE tagged MBP. This synthesis is
promoted by the Ptac promoter, which is a strong promoter. The expression of the fusion
protein was checked with the help of BL21-Gold and CodonPlus. This was mainly done to
eliminate the codon bias issue that might obstruct the expression of the fusion protein in
E.coli hosts. The BL21-Gold cells boost the expression of protein by supplying extra copies
of target specific tRNA genes which are not common in the E. coli genome. In BL21 Gold
and Codon Plus the expressions of the fusion protein was checked. Cells were cultured in
minimal media and the expression of the protein was checked at varying temperatures. The
fusion protein produced was purified by single step affinity purification with a resin having

8SilE: Its characteristics and role in bacterial silver resistance
higher affinity for maltose binding protein (MBP) (Zhao, Li & Liang, 2013). The column
used for purification was packed with amylose resin. Amylose is highly specific in binding to
maltose bound fusion protein in a single step (Kimple, Brill, & Pasker, 2013). The column
was regenerated for multiple use. The fusion protein was subjected to a protease, Factor Xa.
This protease chops the protein after amino acid arginine. The most common cleavage site for
this protease enzyme is Aspartate-Glycine-Arginine (Asp-Gly-Arg) (Chen et al., 2013). The
protein of interest SilE, was collected in the flow-through. The flow through was subjected to
protein dialysis. The purity of the desired protein SilE was checked by the gel filtration
column. The expression and purification using 15N ammonium chloride was repeated to
produce labelled protein for NMR.
Fig 2: Gel image of SilE purification
The gel electrophoresis was carried out after the purification process to ensure the
presence of the SilE protein. The molecular weight of the desired protein is 13kDa. The
presence of the protein is confirmed in Elution 1, 2 and 3 since the high intensity of the bands
in the three lanes are evident from the gel image. The elution 3 is purer in comparison to
elution 1 and 2. The other elution fractions namely elution 4, 5, 6, flow through and wash
13kDa
higher affinity for maltose binding protein (MBP) (Zhao, Li & Liang, 2013). The column
used for purification was packed with amylose resin. Amylose is highly specific in binding to
maltose bound fusion protein in a single step (Kimple, Brill, & Pasker, 2013). The column
was regenerated for multiple use. The fusion protein was subjected to a protease, Factor Xa.
This protease chops the protein after amino acid arginine. The most common cleavage site for
this protease enzyme is Aspartate-Glycine-Arginine (Asp-Gly-Arg) (Chen et al., 2013). The
protein of interest SilE, was collected in the flow-through. The flow through was subjected to
protein dialysis. The purity of the desired protein SilE was checked by the gel filtration
column. The expression and purification using 15N ammonium chloride was repeated to
produce labelled protein for NMR.
Fig 2: Gel image of SilE purification
The gel electrophoresis was carried out after the purification process to ensure the
presence of the SilE protein. The molecular weight of the desired protein is 13kDa. The
presence of the protein is confirmed in Elution 1, 2 and 3 since the high intensity of the bands
in the three lanes are evident from the gel image. The elution 3 is purer in comparison to
elution 1 and 2. The other elution fractions namely elution 4, 5, 6, flow through and wash
13kDa
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

9SilE: Its characteristics and role in bacterial silver resistance
fractions also failed to show intense desired band and hence these fractions were not
considered. So, the elution 3 was concentrated separately since it was purer, having an
intense single band whereas elution 1 and elution 2 was pooled together and concentrated as
multiple bands were observed along with the desired band. The following gel shows the
result of concentrating the respective samples.
Fig 3: Gel image of SilE after concentration
The concentrated samples were again run in the agarose gel to check the presence of
any other junk bands. But the elution fractions gave a clear intense band after concentration
indicating the presence of the pure protein.
Characterisation of SilE based on the silver induced folding
The second objective of this research was to emphasize on the characterisation of the
silver induced folding of the SilE protein.
Previous studies show that SilE undergoes silver-induced folding. It was important to
understand whether the protein extracted is folded or unfolded. This was done by using
Heteronuclear Single Quantum Coherence (HSQC) is a technique frequently exploited in the
NMR spectroscopy of organic molecules being specific to protein NMR (Foroozandeh et al.,
2014). In this spectroscopy each amino acid of the protein is represented as a single peak. The
chemical shifts in the peak and effect in the presence of excess silver was monitored and
13kDa
fractions also failed to show intense desired band and hence these fractions were not
considered. So, the elution 3 was concentrated separately since it was purer, having an
intense single band whereas elution 1 and elution 2 was pooled together and concentrated as
multiple bands were observed along with the desired band. The following gel shows the
result of concentrating the respective samples.
Fig 3: Gel image of SilE after concentration
The concentrated samples were again run in the agarose gel to check the presence of
any other junk bands. But the elution fractions gave a clear intense band after concentration
indicating the presence of the pure protein.
Characterisation of SilE based on the silver induced folding
The second objective of this research was to emphasize on the characterisation of the
silver induced folding of the SilE protein.
Previous studies show that SilE undergoes silver-induced folding. It was important to
understand whether the protein extracted is folded or unfolded. This was done by using
Heteronuclear Single Quantum Coherence (HSQC) is a technique frequently exploited in the
NMR spectroscopy of organic molecules being specific to protein NMR (Foroozandeh et al.,
2014). In this spectroscopy each amino acid of the protein is represented as a single peak. The
chemical shifts in the peak and effect in the presence of excess silver was monitored and
13kDa
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
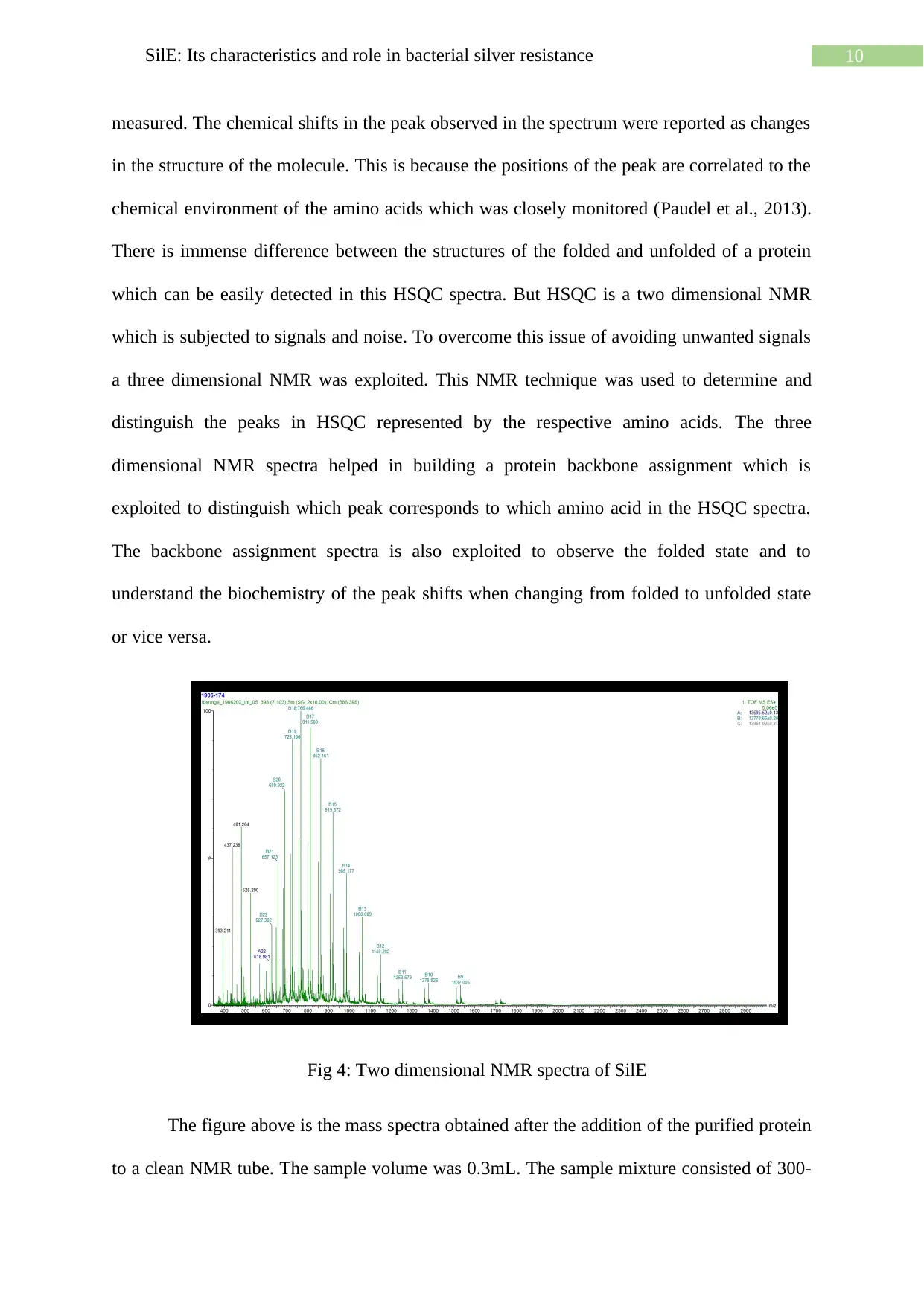
10SilE: Its characteristics and role in bacterial silver resistance
measured. The chemical shifts in the peak observed in the spectrum were reported as changes
in the structure of the molecule. This is because the positions of the peak are correlated to the
chemical environment of the amino acids which was closely monitored (Paudel et al., 2013).
There is immense difference between the structures of the folded and unfolded of a protein
which can be easily detected in this HSQC spectra. But HSQC is a two dimensional NMR
which is subjected to signals and noise. To overcome this issue of avoiding unwanted signals
a three dimensional NMR was exploited. This NMR technique was used to determine and
distinguish the peaks in HSQC represented by the respective amino acids. The three
dimensional NMR spectra helped in building a protein backbone assignment which is
exploited to distinguish which peak corresponds to which amino acid in the HSQC spectra.
The backbone assignment spectra is also exploited to observe the folded state and to
understand the biochemistry of the peak shifts when changing from folded to unfolded state
or vice versa.
Fig 4: Two dimensional NMR spectra of SilE
The figure above is the mass spectra obtained after the addition of the purified protein
to a clean NMR tube. The sample volume was 0.3mL. The sample mixture consisted of 300-
measured. The chemical shifts in the peak observed in the spectrum were reported as changes
in the structure of the molecule. This is because the positions of the peak are correlated to the
chemical environment of the amino acids which was closely monitored (Paudel et al., 2013).
There is immense difference between the structures of the folded and unfolded of a protein
which can be easily detected in this HSQC spectra. But HSQC is a two dimensional NMR
which is subjected to signals and noise. To overcome this issue of avoiding unwanted signals
a three dimensional NMR was exploited. This NMR technique was used to determine and
distinguish the peaks in HSQC represented by the respective amino acids. The three
dimensional NMR spectra helped in building a protein backbone assignment which is
exploited to distinguish which peak corresponds to which amino acid in the HSQC spectra.
The backbone assignment spectra is also exploited to observe the folded state and to
understand the biochemistry of the peak shifts when changing from folded to unfolded state
or vice versa.
Fig 4: Two dimensional NMR spectra of SilE
The figure above is the mass spectra obtained after the addition of the purified protein
to a clean NMR tube. The sample volume was 0.3mL. The sample mixture consisted of 300-

11SilE: Its characteristics and role in bacterial silver resistance
350μM of 15N labelled silver bound SilE at a pH of 7.5 in 10mM of HEPES buffer and 20mM
NaF (sodium fluoride) and the volume was made up with 10% D2O. The sample was placed
inside the spectrometer in a homogeneous magnetic field. 15N/1H Heteronuclear Single
Quantum Coherence (HSQC) spectra was obtained at a temperature of 25 C⁰ (Asiani et al.,
2016) and the data was collected accordingly. The extra noise and signals obtained in the two
dimensional NMR and to further study the details of the induced folding of SilE in the
presence of silver led to the exploitation of Nuclear Overhauser Effects, three dimensional
NMR.
Acquiring the structural information of the silver-bound folded SilE
Nuclear Overhauser Effects (NOE) is an NMR measurement exploited to calculate the
distance between protons in the folded structure of a protein (Emsley, Feeney & Sutcliffe,
2013). This technique was exploited in this article to understand the folding of the SilE
protein in the presence of silver ions. The residual dipolar coupling (RDC) is exploited to
determine the protein structure (Akitt & Mann, 2017). RDC is the measurement that provides
information on the orientation of the residues involved in the magnetic interaction (Khoo,
Singer & Cowburn, 2017). Paramagnetic relaxation enhancements (PREs) is the
measurement of distance in restraints exploiting the site-specific attachment of the
paramagnetic spin labels to proteins (Sjodt & Clubb, 2017). The protein is subjected to
cysteine mutation and tagged with MTSL. This is done to detect the differences observed in
the height of the measured peaks exploiting the presence of the organosulphur compound,
MTSL (Jackman & Sternhell, 2013). It measures the distance between the tag itself and the
amino acids in its vicinity. MTSL was used as a nitroxide spin label. The MTSL is attached
to the protein via thiol groups and hence, it exploits the thiol of the amino acid cysteine. Site
directed spin labelling occurs due to the hetero disulfide bond formed.
350μM of 15N labelled silver bound SilE at a pH of 7.5 in 10mM of HEPES buffer and 20mM
NaF (sodium fluoride) and the volume was made up with 10% D2O. The sample was placed
inside the spectrometer in a homogeneous magnetic field. 15N/1H Heteronuclear Single
Quantum Coherence (HSQC) spectra was obtained at a temperature of 25 C⁰ (Asiani et al.,
2016) and the data was collected accordingly. The extra noise and signals obtained in the two
dimensional NMR and to further study the details of the induced folding of SilE in the
presence of silver led to the exploitation of Nuclear Overhauser Effects, three dimensional
NMR.
Acquiring the structural information of the silver-bound folded SilE
Nuclear Overhauser Effects (NOE) is an NMR measurement exploited to calculate the
distance between protons in the folded structure of a protein (Emsley, Feeney & Sutcliffe,
2013). This technique was exploited in this article to understand the folding of the SilE
protein in the presence of silver ions. The residual dipolar coupling (RDC) is exploited to
determine the protein structure (Akitt & Mann, 2017). RDC is the measurement that provides
information on the orientation of the residues involved in the magnetic interaction (Khoo,
Singer & Cowburn, 2017). Paramagnetic relaxation enhancements (PREs) is the
measurement of distance in restraints exploiting the site-specific attachment of the
paramagnetic spin labels to proteins (Sjodt & Clubb, 2017). The protein is subjected to
cysteine mutation and tagged with MTSL. This is done to detect the differences observed in
the height of the measured peaks exploiting the presence of the organosulphur compound,
MTSL (Jackman & Sternhell, 2013). It measures the distance between the tag itself and the
amino acids in its vicinity. MTSL was used as a nitroxide spin label. The MTSL is attached
to the protein via thiol groups and hence, it exploits the thiol of the amino acid cysteine. Site
directed spin labelling occurs due to the hetero disulfide bond formed.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 21
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2025 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.

