Applying Cox-PH Model for Survival Analysis in R Statistics
VerifiedAdded on 2023/05/29
|6
|876
|350
Homework Assignment
AI Summary
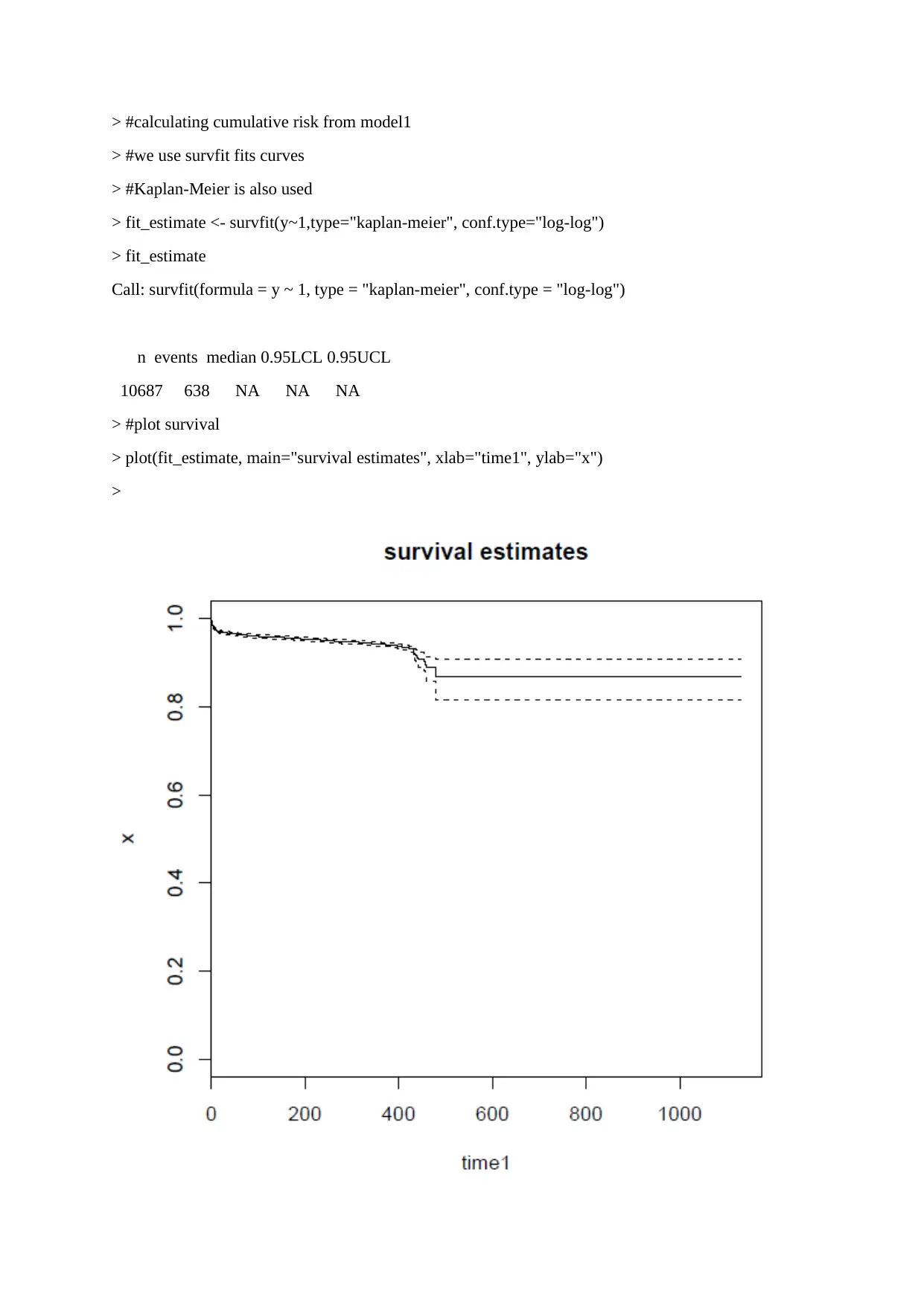
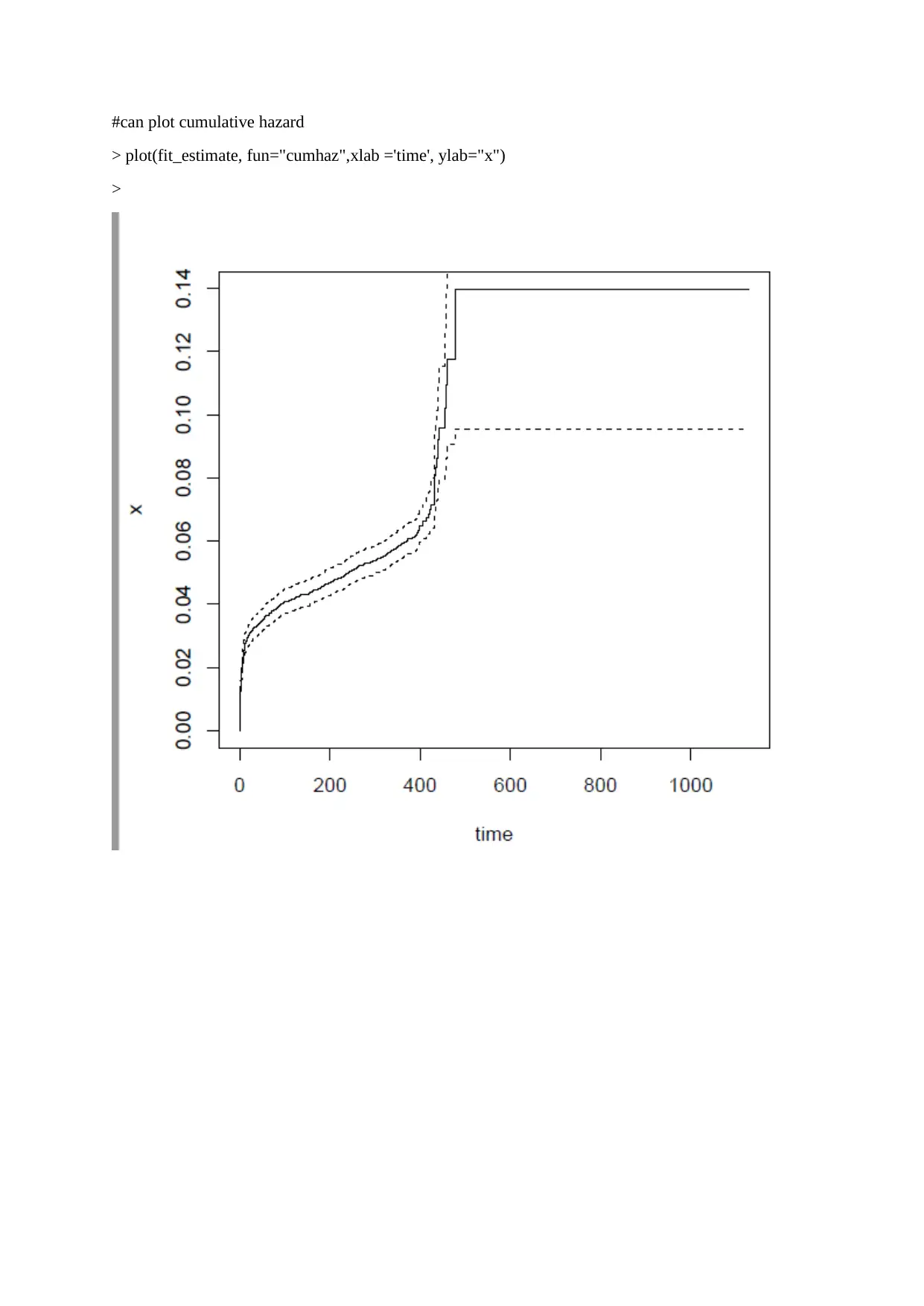
This assignment provides a detailed solution to a survival analysis problem using R statistics, focusing on the Cox-PH model. The solution includes calculating the expected risk (cumulative incidence) of events based on different conditions using models derived from the provided R code. It further involves calculating the difference between predicted values from two models and fitting a linear regression model using this difference as the outcome. Finally, the assignment tests the squared semi-partial correlations and partial correlations for each variable in the linear regression model, selecting variables that statistically account for more than 1% of the observed variations. The code and results offer a comprehensive approach to analyzing survival data and understanding the impact of various covariates.
1 out of 6











![[object Object]](/_next/static/media/star-bottom.7253800d.svg)