Biochemistry Project: SWI2/SNF2 and TLR5 Proteins Analysis
VerifiedAdded on 2023/06/10
|21
|2484
|280
Project
AI Summary
This bioinformatics project presents a detailed analysis of two key proteins: SWI2/SNF2 and TLR5. The SWI2/SNF2 section focuses on its role as a chromatin-remodeling complex, examining the effects of mutations on ATP hydrolysis and DNA wrapping. It explores the structural features, including helix structures and the impact of amino acid changes on energy release and charge distribution, with implications for protein folding and DNA packaging. The TLR5 section investigates its role in immune responses, specifically its interaction with flagellin and its involvement in osteoclastogenesis and bone loss. The analysis includes hydrogen bonding, protein shape alterations, and conserved amino acid analysis, highlighting how changes in protein structure can affect ligand binding and receptor function, ultimately influencing immune pathways and bone metabolism. The project uses bioinformatics tools to analyze protein structures, mutation effects, and structural changes in the context of their biological functions.

BIOCHEMISTRY PROJECT
Submitted to:
Submitted by:
Submitted to:
Submitted by:
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Assigned protein: SWI2/SNF2 chromatin-remodeling
PDB code: 1Z3l
Introduction
SWI/SNF (SWItch/Sucrose Non-Fermentable) found in both eukaryotes and prokaryotes and
referred as nucleosome remodeling complex. In other words it is a cluster of proteins related to
DNA packaging behavior. It comprises of numerous proteins – products of the SNF and SWI
genes (SWI1, SWI2/SNF2, SWI3, SWI5, SWI6) and many other various polypeptides.it can
disrupt the interaction of DNA and histone in ATP dependent manner due the presence of DNA
stimulated ATPase activity.
SWI2/SNF2 proteins are multi domain that can interact with more than one protein partners and
inherently flexible molecules that strangle the crystallization. SWl2/SNF2 chromatin remodeling
proteins (DNA repair enzymes) intercede the mobilization of DNA associated proteins and
nucleosome. By using ATP-Hydrolysis, these molecular motors transport the duplex DNA along
a helicase‐like domain. Binding of SWI2/SNF2 enzymes to the DNA duplex in a structure
independent manner that provides an attribute of challenging obstacle for specific protein
crystallization. Conservation of ATPase domain in all the SWI2/SNF2 enzymes representing the
conserved ATP-driven processes on DNA among various family members (McKay and
Caruthers, 2002, Peterson and Richmond, 1996). Interaction between SWI2/SNF2 comlex and
chromatin permits the transcription factor binding and as a result promote transcription.
Chromodomains are also of the prime importance because of their activity as the removal of
chromodomains improve the ATPase activity.
PDB code: 1Z3l
Introduction
SWI/SNF (SWItch/Sucrose Non-Fermentable) found in both eukaryotes and prokaryotes and
referred as nucleosome remodeling complex. In other words it is a cluster of proteins related to
DNA packaging behavior. It comprises of numerous proteins – products of the SNF and SWI
genes (SWI1, SWI2/SNF2, SWI3, SWI5, SWI6) and many other various polypeptides.it can
disrupt the interaction of DNA and histone in ATP dependent manner due the presence of DNA
stimulated ATPase activity.
SWI2/SNF2 proteins are multi domain that can interact with more than one protein partners and
inherently flexible molecules that strangle the crystallization. SWl2/SNF2 chromatin remodeling
proteins (DNA repair enzymes) intercede the mobilization of DNA associated proteins and
nucleosome. By using ATP-Hydrolysis, these molecular motors transport the duplex DNA along
a helicase‐like domain. Binding of SWI2/SNF2 enzymes to the DNA duplex in a structure
independent manner that provides an attribute of challenging obstacle for specific protein
crystallization. Conservation of ATPase domain in all the SWI2/SNF2 enzymes representing the
conserved ATP-driven processes on DNA among various family members (McKay and
Caruthers, 2002, Peterson and Richmond, 1996). Interaction between SWI2/SNF2 comlex and
chromatin permits the transcription factor binding and as a result promote transcription.
Chromodomains are also of the prime importance because of their activity as the removal of
chromodomains improve the ATPase activity.

Initially DNA binds to the high affinity DBD and conformational changes occur in the active
domain in the presence of ATP. These conformational changes allow dsDNA to move in the
active site of SWI2/SNF2 in the same pattern as ssDNA transported in the helicase active site.
Swi2/Snf2 domains have tracked along the minor groove of dsDNA by using ATP (Durr et al.
2005, Zhang. et al, 2006)
Results
1. Mutation in Helix
SWI2/SNF2 chromatin-remodeling protein consist of three helix structures. Only one of the helix
contains Glutamic acid (GLU9) and Glutamine (GLN11) while others only have Glutamine
(GLN29 and GLN55). As it is observed in the previous studies that glutamine can abolish ATP
hydrolysis. So mutation has been done at the site of glutamic acid (GLU9) as shown in the
figure.1, figure.2a and figure.2b.
Fig.1
domain in the presence of ATP. These conformational changes allow dsDNA to move in the
active site of SWI2/SNF2 in the same pattern as ssDNA transported in the helicase active site.
Swi2/Snf2 domains have tracked along the minor groove of dsDNA by using ATP (Durr et al.
2005, Zhang. et al, 2006)
Results
1. Mutation in Helix
SWI2/SNF2 chromatin-remodeling protein consist of three helix structures. Only one of the helix
contains Glutamic acid (GLU9) and Glutamine (GLN11) while others only have Glutamine
(GLN29 and GLN55). As it is observed in the previous studies that glutamine can abolish ATP
hydrolysis. So mutation has been done at the site of glutamic acid (GLU9) as shown in the
figure.1, figure.2a and figure.2b.
Fig.1
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
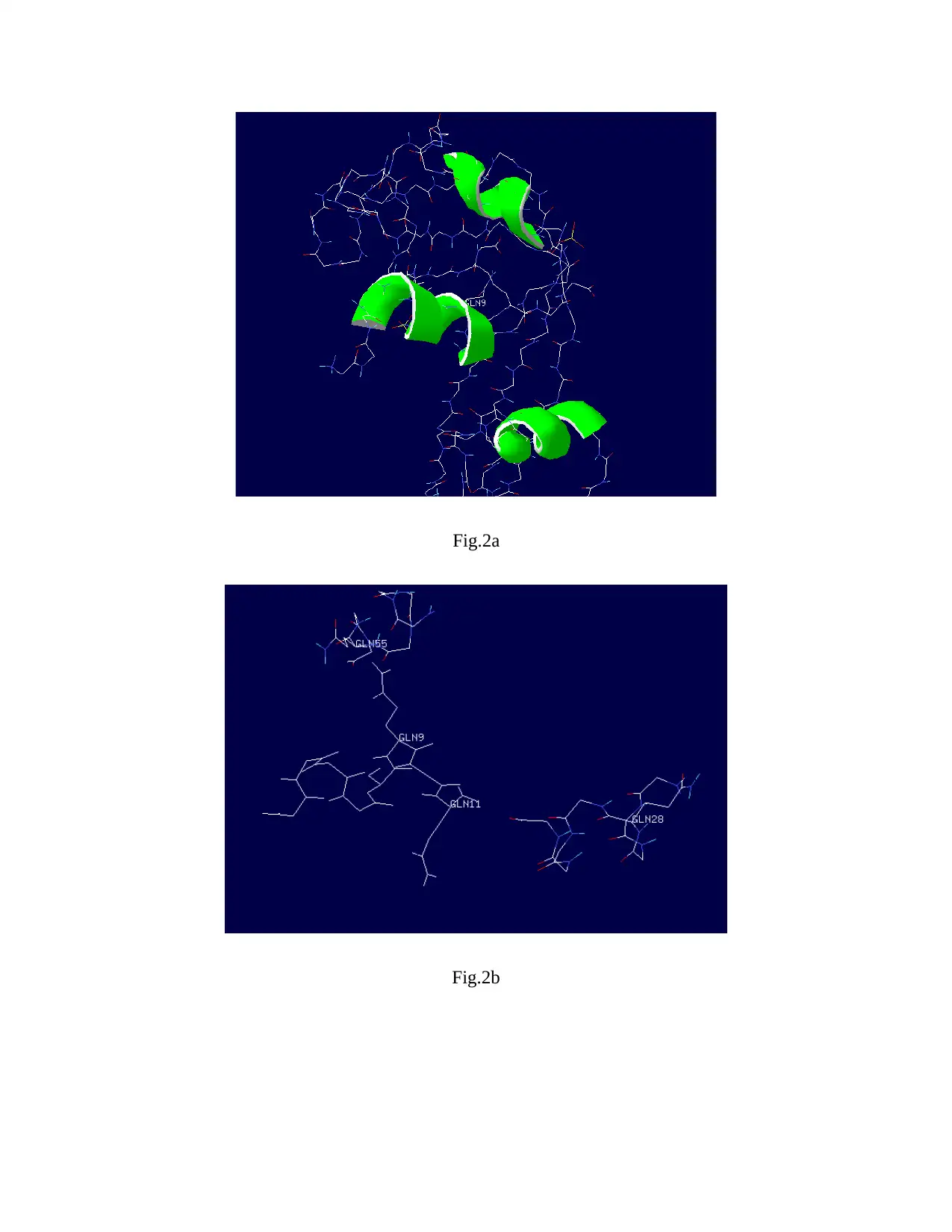
Fig.2a
Fig.2b
Fig.2b
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

2. Energy Difference
The evidence of decrease in ATP hydrolysis can be seen by the total energy of folding of a
protein as in normal SWI2/SNF2 chromatin-remodeling protein, the energy released is E= -
5518.564 KJ/mol while mutated SWI2/SNF2 protein released E= -5626.675 KJ/mol shown in
figure.3 and figure.4. This decrease in hydrolysis due to substitution mutation has no effect on
ATP binding.
Fig.3
Fig.4
3. Charges on sections of the protein
The charge on different amino acid side chain can vary when they are close to highly charged or
non-polar chains. Figure.5 and figure.6 showed the charge on the non-mutated and mutated
section of the SWI2/SNF2 chromatin-remodeling protein. As the side chain of glutamic acid
(GLU9) is not longer than the mutated glutamine (GLN9), the charge distribution on the
glutamine is wider than the glutamic acid. Due to wider charge distribution, the attraction or
repulsion of this mutated site is more as compared to wild type. Like in Arabidopsis the
The evidence of decrease in ATP hydrolysis can be seen by the total energy of folding of a
protein as in normal SWI2/SNF2 chromatin-remodeling protein, the energy released is E= -
5518.564 KJ/mol while mutated SWI2/SNF2 protein released E= -5626.675 KJ/mol shown in
figure.3 and figure.4. This decrease in hydrolysis due to substitution mutation has no effect on
ATP binding.
Fig.3
Fig.4
3. Charges on sections of the protein
The charge on different amino acid side chain can vary when they are close to highly charged or
non-polar chains. Figure.5 and figure.6 showed the charge on the non-mutated and mutated
section of the SWI2/SNF2 chromatin-remodeling protein. As the side chain of glutamic acid
(GLU9) is not longer than the mutated glutamine (GLN9), the charge distribution on the
glutamine is wider than the glutamic acid. Due to wider charge distribution, the attraction or
repulsion of this mutated site is more as compared to wild type. Like in Arabidopsis the
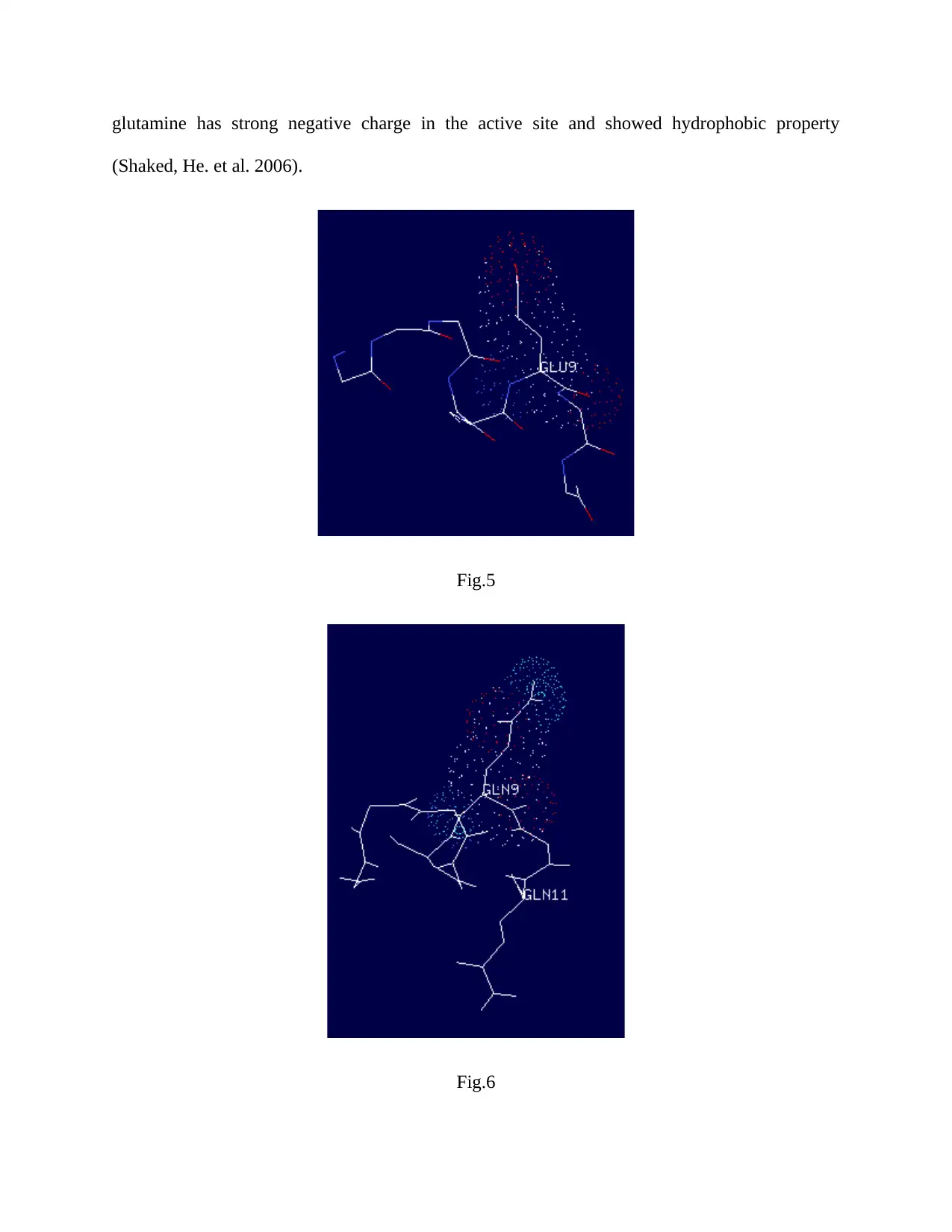
glutamine has strong negative charge in the active site and showed hydrophobic property
(Shaked, He. et al. 2006).
Fig.5
Fig.6
(Shaked, He. et al. 2006).
Fig.5
Fig.6
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

4. Measure angles between key atoms/residues
Side chain angle showed difference in angle between non-mutated and mutated amino acid. Non-
mutated amino acid (GLU9) showed angle of 129.4֯ (Fig.7), while mutated amino acid (GLN9)
showed 130.07֯ angle (Fig.8). The Shape of the protein is governed by the interaction of amino
acid side chains. Although this mutation showed little difference in the angle of the side chain
but the angle of the side chains plays important role in the folding of the proteins as various
amino acid side chains interact with each other by establishing ionic and hydrogen bonds. By
increasing the Glutamine side chain angle we can provide a chance of hydrogen bonding of the
Helix with the other amino acid side chains that can reduce the ATP hydrolysis without
disturbing its binding property. This bonding as a result cause different type of DNA packaging.
Fig.7
Side chain angle showed difference in angle between non-mutated and mutated amino acid. Non-
mutated amino acid (GLU9) showed angle of 129.4֯ (Fig.7), while mutated amino acid (GLN9)
showed 130.07֯ angle (Fig.8). The Shape of the protein is governed by the interaction of amino
acid side chains. Although this mutation showed little difference in the angle of the side chain
but the angle of the side chains plays important role in the folding of the proteins as various
amino acid side chains interact with each other by establishing ionic and hydrogen bonds. By
increasing the Glutamine side chain angle we can provide a chance of hydrogen bonding of the
Helix with the other amino acid side chains that can reduce the ATP hydrolysis without
disturbing its binding property. This bonding as a result cause different type of DNA packaging.
Fig.7
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
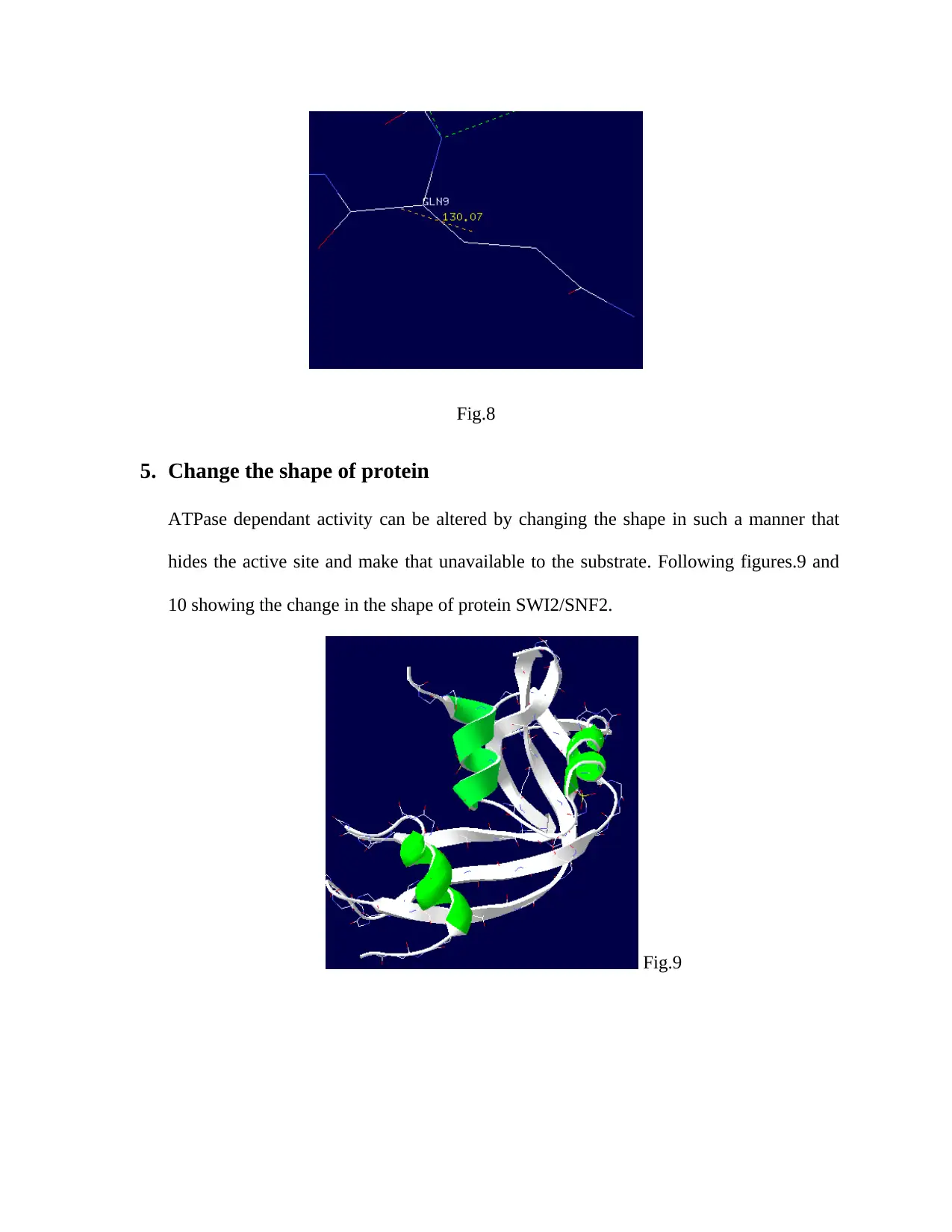
Fig.8
5. Change the shape of protein
ATPase dependant activity can be altered by changing the shape in such a manner that
hides the active site and make that unavailable to the substrate. Following figures.9 and
10 showing the change in the shape of protein SWI2/SNF2.
Fig.9
5. Change the shape of protein
ATPase dependant activity can be altered by changing the shape in such a manner that
hides the active site and make that unavailable to the substrate. Following figures.9 and
10 showing the change in the shape of protein SWI2/SNF2.
Fig.9
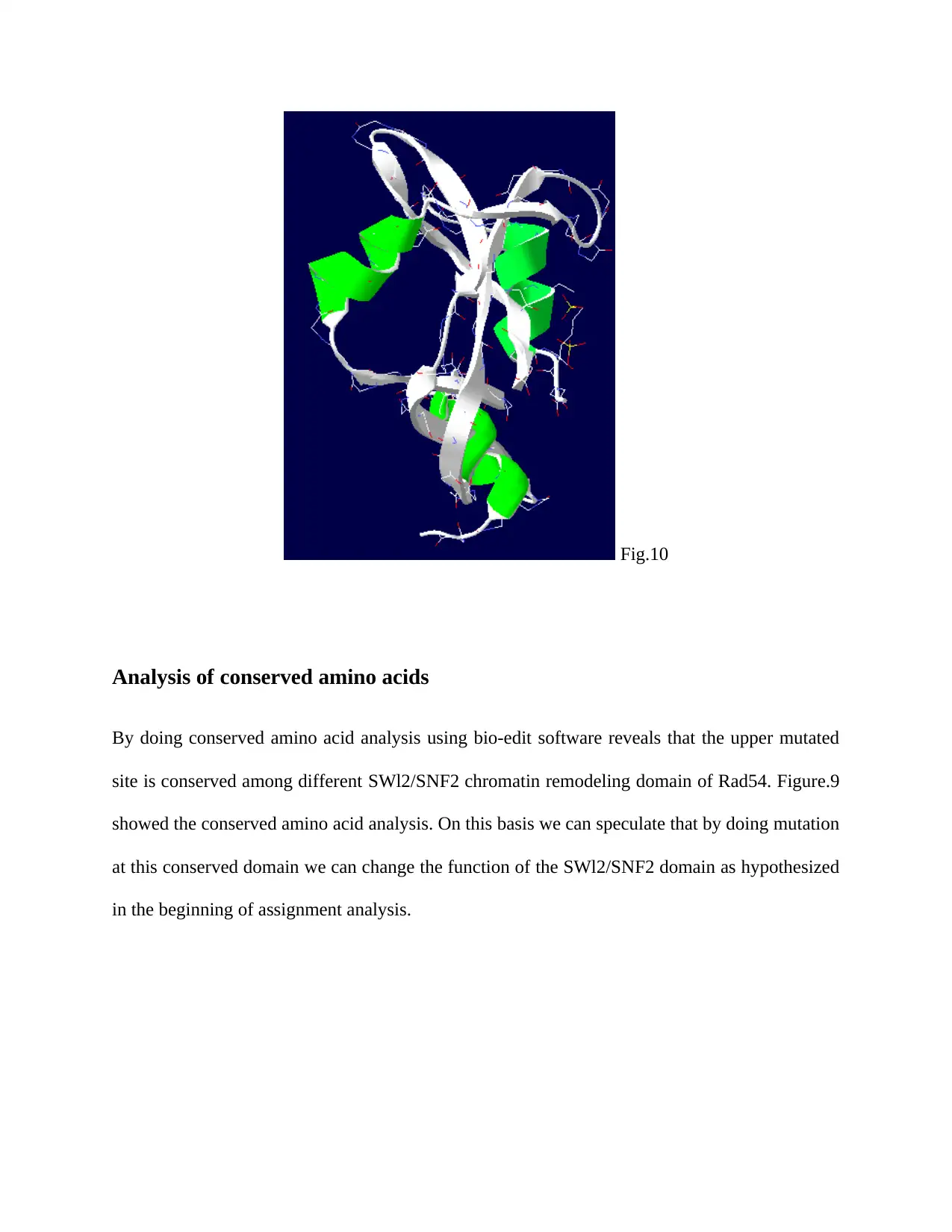
Fig.10
Analysis of conserved amino acids
By doing conserved amino acid analysis using bio-edit software reveals that the upper mutated
site is conserved among different SWl2/SNF2 chromatin remodeling domain of Rad54. Figure.9
showed the conserved amino acid analysis. On this basis we can speculate that by doing mutation
at this conserved domain we can change the function of the SWl2/SNF2 domain as hypothesized
in the beginning of assignment analysis.
Analysis of conserved amino acids
By doing conserved amino acid analysis using bio-edit software reveals that the upper mutated
site is conserved among different SWl2/SNF2 chromatin remodeling domain of Rad54. Figure.9
showed the conserved amino acid analysis. On this basis we can speculate that by doing mutation
at this conserved domain we can change the function of the SWl2/SNF2 domain as hypothesized
in the beginning of assignment analysis.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Conclusion
SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 contain two chains having three
helix in the crystal structure. Only one of the helix contain glutamic acid that lies in conserved
domain among different organisms. While wrapping up DNA wild protein release -5518.564
KJ/mol energy in the form of ATP while after mutating this glutamic acid into glutamine the
energy released was -5626.675 KJ/mol. This result showed that glutamine hinders the ATP
hydrolysis resulted in less energy release during DNA wrapping. The charge on the side chain of
the mutated site was widespread than the non-mutated site indicating the possibility of strong
interaction. More over the angle is more in mutated side chain as compared to non-mutated.
This angle difference cause hydrogen bonding between mutated side chain (glutamine) and other
side chains which is way stronger than the wild type (glutamic acid) salt bridges.
According to these findings, by mutating Glutamic acid into glutamine in the helix of
SWI2/SNF2 chromatin domain ATPase dependent activity can be altered. This change can cause
positive or negative change in the wrapping of DNA and ultimately change the way of specific
protein crystallization.
SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 contain two chains having three
helix in the crystal structure. Only one of the helix contain glutamic acid that lies in conserved
domain among different organisms. While wrapping up DNA wild protein release -5518.564
KJ/mol energy in the form of ATP while after mutating this glutamic acid into glutamine the
energy released was -5626.675 KJ/mol. This result showed that glutamine hinders the ATP
hydrolysis resulted in less energy release during DNA wrapping. The charge on the side chain of
the mutated site was widespread than the non-mutated site indicating the possibility of strong
interaction. More over the angle is more in mutated side chain as compared to non-mutated.
This angle difference cause hydrogen bonding between mutated side chain (glutamine) and other
side chains which is way stronger than the wild type (glutamic acid) salt bridges.
According to these findings, by mutating Glutamic acid into glutamine in the helix of
SWI2/SNF2 chromatin domain ATPase dependent activity can be altered. This change can cause
positive or negative change in the wrapping of DNA and ultimately change the way of specific
protein crystallization.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

References:
(1) Durr, H., Korner, C., Muller, M., Hickmann, V., and Hopfner, K. P. (2005). X‐ray
structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with
DNA. Cell 121, 363–373.
(2) Caruthers, J.M. and McKay, D.B. Helicase structure and mechanism. Curr. Opin. Struct.
Biol. 2002; 12: 123–133
(3) Richmond, E. and Peterson, C.L. Functional analysis of the DNA-stimulated ATPase
domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996; 24: 3685–3692
(4) Y. Zhang, C.L. Smith, A. Saha, S.W. Grill, S. Mihardja, S.B. Smith, B.R. Cairns, C.L.
Peterson, C. Bustamante. DNA translocation and loop formation mechanism of
chromatin remodeling by SWI/SNF and RSC Mol Cell, 24 (2006), pp. 559-568
(5) Harald, DU., and Karl, P. Structure–Function Analysis of SWI2/SNF2 Enzymes. 2006.
Method in enzymology, VOL. 409
(6) Shaked, He., Avivi-Ragolsky., No., and Levy., A. Involvement of the Arabidopsis
SWI2/SNF2 Chromatin Remodeling Gene Family in DNA Damage Response and
Recombination. GENETICS June 1, 2006 vol. 173 no. 2 985-994
Introduction
Toll-like receptors (TLRs) are a cluster of proteins that play pivotal role in immune system to
recognize pathogens. These receptors start intracellular signal transduction by recruiting and
activating the adapter molecules after recognition of microbial molecules. In response to these
adapters e.g. TRAF6, different TFs (transcription factors) e.g. NFKB and MAPKs (Mitogen
(1) Durr, H., Korner, C., Muller, M., Hickmann, V., and Hopfner, K. P. (2005). X‐ray
structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with
DNA. Cell 121, 363–373.
(2) Caruthers, J.M. and McKay, D.B. Helicase structure and mechanism. Curr. Opin. Struct.
Biol. 2002; 12: 123–133
(3) Richmond, E. and Peterson, C.L. Functional analysis of the DNA-stimulated ATPase
domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996; 24: 3685–3692
(4) Y. Zhang, C.L. Smith, A. Saha, S.W. Grill, S. Mihardja, S.B. Smith, B.R. Cairns, C.L.
Peterson, C. Bustamante. DNA translocation and loop formation mechanism of
chromatin remodeling by SWI/SNF and RSC Mol Cell, 24 (2006), pp. 559-568
(5) Harald, DU., and Karl, P. Structure–Function Analysis of SWI2/SNF2 Enzymes. 2006.
Method in enzymology, VOL. 409
(6) Shaked, He., Avivi-Ragolsky., No., and Levy., A. Involvement of the Arabidopsis
SWI2/SNF2 Chromatin Remodeling Gene Family in DNA Damage Response and
Recombination. GENETICS June 1, 2006 vol. 173 no. 2 985-994
Introduction
Toll-like receptors (TLRs) are a cluster of proteins that play pivotal role in immune system to
recognize pathogens. These receptors start intracellular signal transduction by recruiting and
activating the adapter molecules after recognition of microbial molecules. In response to these
adapters e.g. TRAF6, different TFs (transcription factors) e.g. NFKB and MAPKs (Mitogen

activated protein kinases) activated which then control the regulation of many immune
responsive and inflammatory genes. Among different TLRs, TLR5 recognizes the flagellin
stimulation in response to which NFKB activated in cell lines. Zebrafish cell lines can be used to
unravel the different signaling mechanisms and events in response to pathogen recognition.
Zebrafish appear to be tremendously different from than human but in fact about 70% genes of
human are found in zebrafish. Many critical pathways and different genes which are necessary to
grow and proper function of different organs are highly conserved between them. Among TLRs,
TLR5 is known to distinguish flagellin among different invading bacteria. Different studies
showed the involvement of TLRs in many diseases including inflammatory bowel disease (1).
Different recent studies indicating the relation of production of TLR5 with the bone loss and
osteoclastogenesis (2). Moreover, the abnormal functioning of TLR5 causes the ovarian, cervical
and gastric cancers (3,4).
The interaction between flagellin and TLR5 can cause different responses in different type of
cells. Osteoclastogenesis and bone loss is the one of the best example for better understanding of
TLR5 functioning. Rheumatoid arthritis patients contain flagellin in their synovial fluid that
activates TLR5, in response to which RANKL (receptor activator of NFkB ligands) activated.
Due to activation of these ligands the expression of osteoclastic genes enhanced. This increased
expression of genes cause bone loss and osteoclast formation.
Analysis Results:
Figure.1 is showing four chains of different colors. Among these four chains, red and green
chains are of TLR5 while dark blue and light blue colored chains are of Variable Lymphocyte
Receptor 2.
responsive and inflammatory genes. Among different TLRs, TLR5 recognizes the flagellin
stimulation in response to which NFKB activated in cell lines. Zebrafish cell lines can be used to
unravel the different signaling mechanisms and events in response to pathogen recognition.
Zebrafish appear to be tremendously different from than human but in fact about 70% genes of
human are found in zebrafish. Many critical pathways and different genes which are necessary to
grow and proper function of different organs are highly conserved between them. Among TLRs,
TLR5 is known to distinguish flagellin among different invading bacteria. Different studies
showed the involvement of TLRs in many diseases including inflammatory bowel disease (1).
Different recent studies indicating the relation of production of TLR5 with the bone loss and
osteoclastogenesis (2). Moreover, the abnormal functioning of TLR5 causes the ovarian, cervical
and gastric cancers (3,4).
The interaction between flagellin and TLR5 can cause different responses in different type of
cells. Osteoclastogenesis and bone loss is the one of the best example for better understanding of
TLR5 functioning. Rheumatoid arthritis patients contain flagellin in their synovial fluid that
activates TLR5, in response to which RANKL (receptor activator of NFkB ligands) activated.
Due to activation of these ligands the expression of osteoclastic genes enhanced. This increased
expression of genes cause bone loss and osteoclast formation.
Analysis Results:
Figure.1 is showing four chains of different colors. Among these four chains, red and green
chains are of TLR5 while dark blue and light blue colored chains are of Variable Lymphocyte
Receptor 2.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 21
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.
