Comprehensive Study on Polypeptide Synthesis and Meiosis in Biology
VerifiedAdded on 2022/09/01
|13
|988
|20
Homework Assignment
AI Summary
This biology assignment comprehensively examines polypeptide synthesis and meiosis. Part A delves into transcription, detailing the roles of RNA polymerase and the process of adding nucleotides. It explores RNA processing, including capping, polyA tail addition, and splicing. Translation is covered with a focus on nucleotide and protein sequences. The assignment also explores protein folding, the physical process by which a protein acquires its functional 3D structure, and the different levels of protein folding. Part B analyzes the cell cycle during Meiosis I and II, along with mitosis, and explains crossing over, independent assortment, random segregation, and the formation of haploid gametes. The assignment references several scientific publications to support the findings and analysis.

POLYPEPTIDE SYNTHESIS AND MEIOSIS
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

PART A
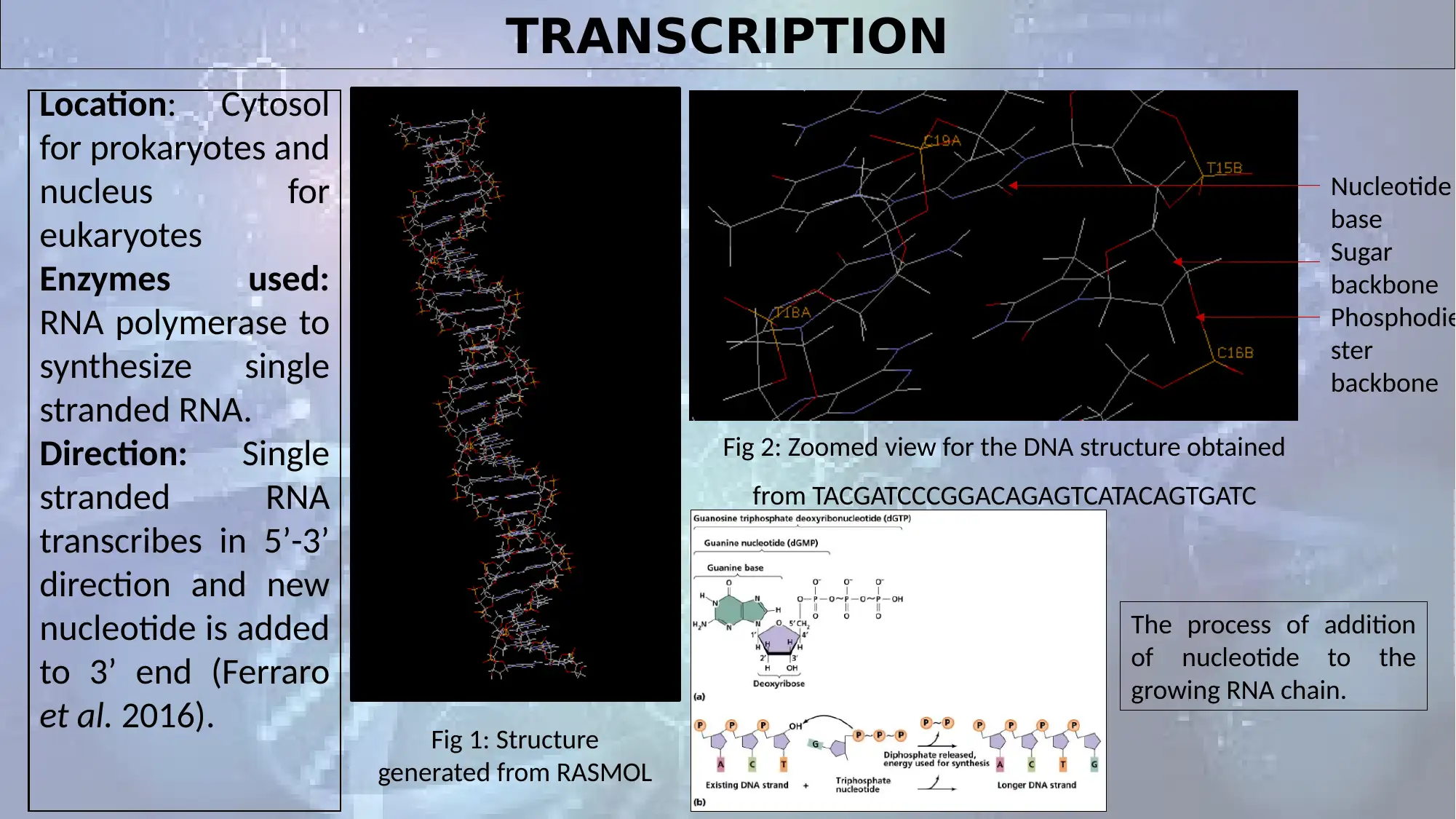
TRANSCRIPTION
Location: Cytosol
for prokaryotes and
nucleus for
eukaryotes
Enzymes used:
RNA polymerase to
synthesize single
stranded RNA.
Direction: Single
stranded RNA
transcribes in 5’-3’
direction and new
nucleotide is added
to 3’ end (Ferraro
et al. 2016). Fig 1: Structure
generated from RASMOL
Nucleotide
base
Sugar
backbone
Phosphodie
ster
backbone
Fig 2: Zoomed view for the DNA structure obtained
from TACGATCCCGGACAGAGTCATACAGTGATC
The process of addition
of nucleotide to the
growing RNA chain.
Location: Cytosol
for prokaryotes and
nucleus for
eukaryotes
Enzymes used:
RNA polymerase to
synthesize single
stranded RNA.
Direction: Single
stranded RNA
transcribes in 5’-3’
direction and new
nucleotide is added
to 3’ end (Ferraro
et al. 2016). Fig 1: Structure
generated from RASMOL
Nucleotide
base
Sugar
backbone
Phosphodie
ster
backbone
Fig 2: Zoomed view for the DNA structure obtained
from TACGATCCCGGACAGAGTCATACAGTGATC
The process of addition
of nucleotide to the
growing RNA chain.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
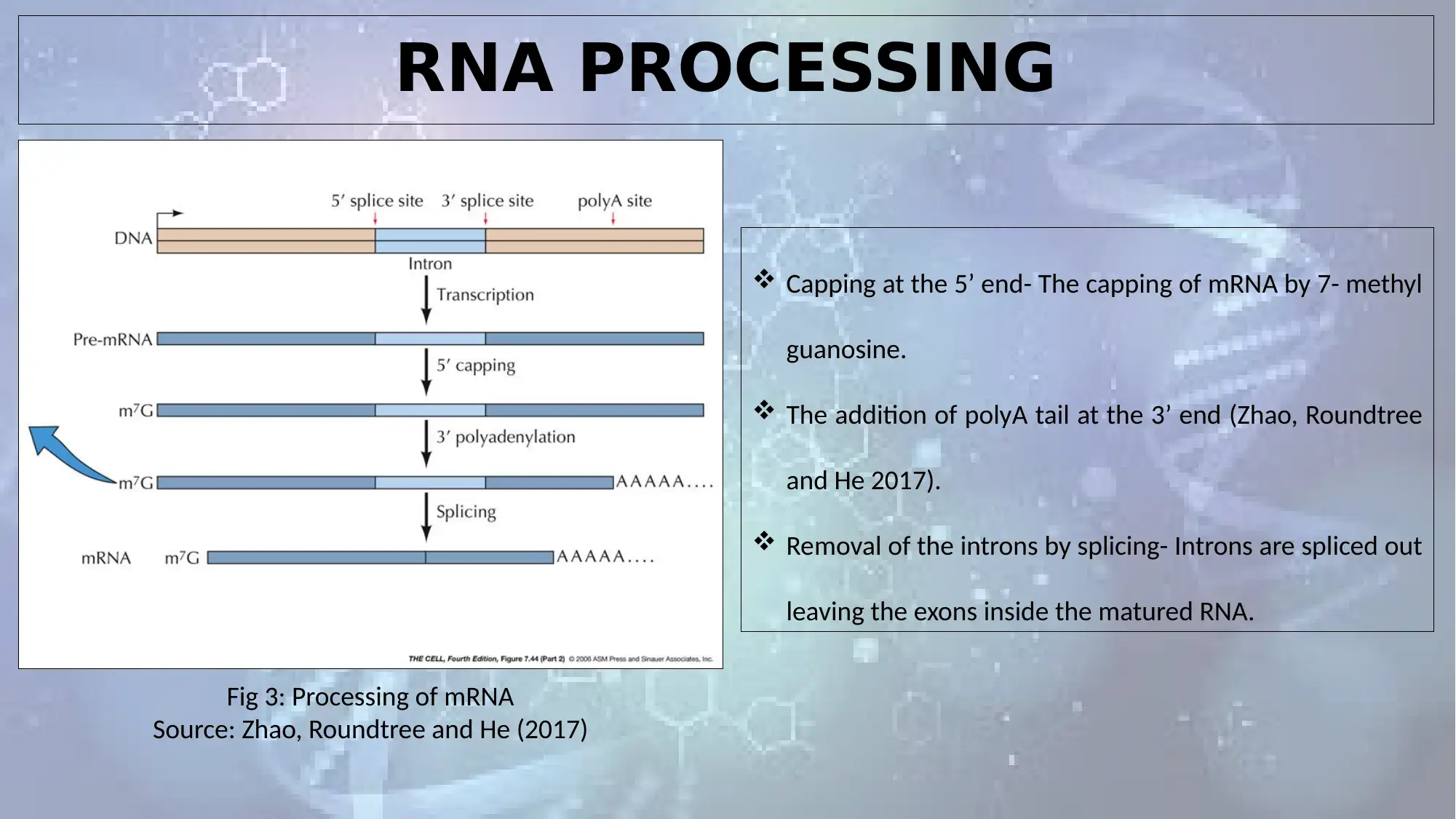
RNA PROCESSING
Fig 3: Processing of mRNA
Source: Zhao, Roundtree and He (2017)
Capping at the 5’ end- The capping of mRNA by 7- methyl
guanosine.
The addition of polyA tail at the 3’ end (Zhao, Roundtree
and He 2017).
Removal of the introns by splicing- Introns are spliced out
leaving the exons inside the matured RNA.
Fig 3: Processing of mRNA
Source: Zhao, Roundtree and He (2017)
Capping at the 5’ end- The capping of mRNA by 7- methyl
guanosine.
The addition of polyA tail at the 3’ end (Zhao, Roundtree
and He 2017).
Removal of the introns by splicing- Introns are spliced out
leaving the exons inside the matured RNA.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
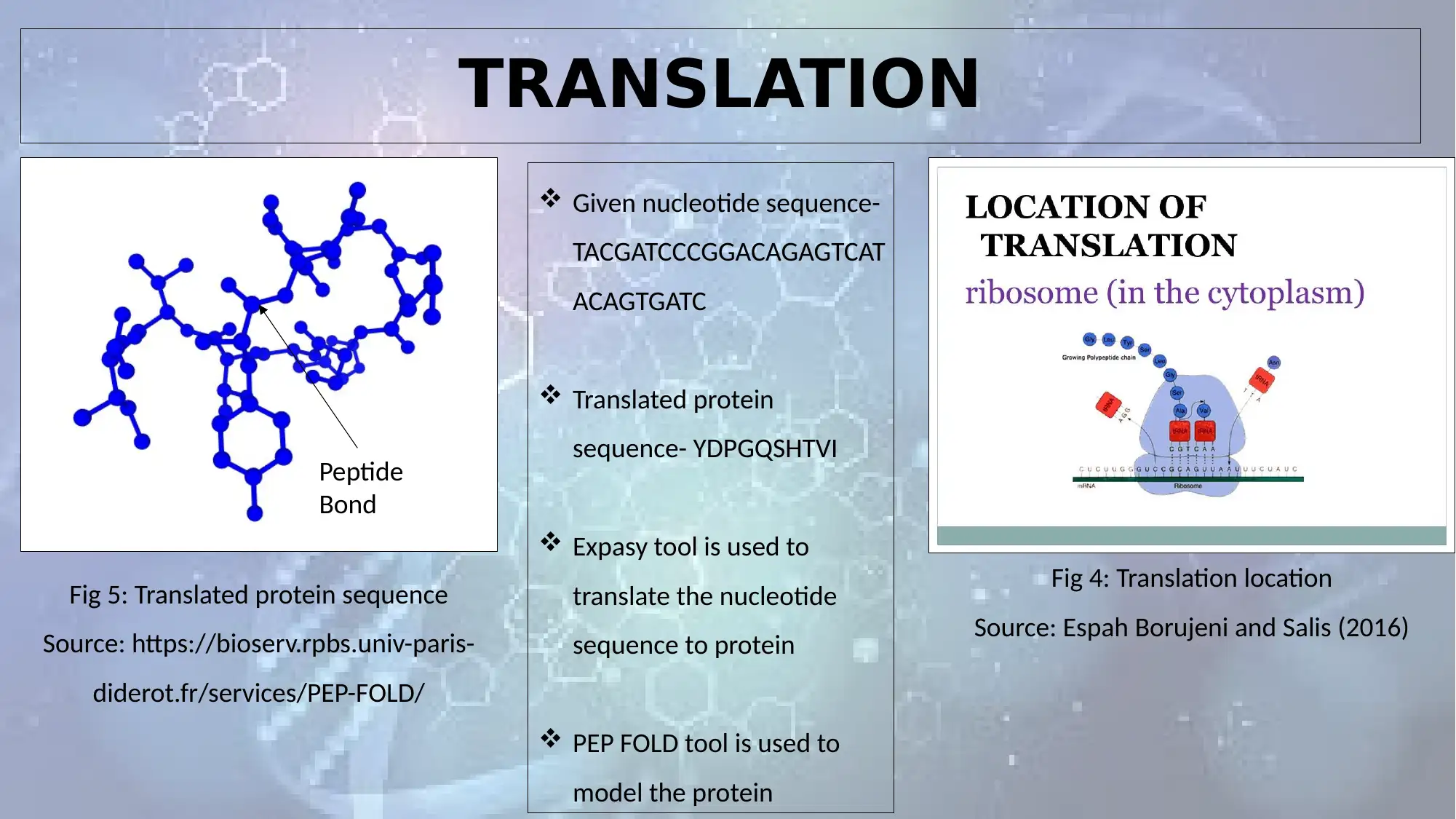
TRANSLATION
Fig 4: Translation location
Source: Espah Borujeni and Salis (2016)
Given nucleotide sequence-
TACGATCCCGGACAGAGTCAT
ACAGTGATC
Translated protein
sequence- YDPGQSHTVI
Expasy tool is used to
translate the nucleotide
sequence to protein
PEP FOLD tool is used to
model the protein
Fig 5: Translated protein sequence
Source: https://bioserv.rpbs.univ-paris-
diderot.fr/services/PEP-FOLD/
Peptide
Bond
Fig 4: Translation location
Source: Espah Borujeni and Salis (2016)
Given nucleotide sequence-
TACGATCCCGGACAGAGTCAT
ACAGTGATC
Translated protein
sequence- YDPGQSHTVI
Expasy tool is used to
translate the nucleotide
sequence to protein
PEP FOLD tool is used to
model the protein
Fig 5: Translated protein sequence
Source: https://bioserv.rpbs.univ-paris-
diderot.fr/services/PEP-FOLD/
Peptide
Bond
PROTEIN FOLDING
The physical process by which a chain of protein acquires its functional form (3 dimensional native structure) (Englander
and Mayne 2017).
While protein folding is performed several residues are deleted and several resides are added to the original structure
(ribosylation, glycosylation, acylation etc.).
This folding process is carried inside the protein folding complex known as proteasome complex or the chaperone
system.
Both eukaryote and prokaryotic organisms has different types of protein folding complexes.
There are four levels of protein folding: Primary, secondary, tertiary and quarternary structure.
Various types of interactions plays an important role during the protein folding process.
Hydrophobic and Vander walls interactions are the most important interactions playing a significant role in protein
synthesis.
The physical process by which a chain of protein acquires its functional form (3 dimensional native structure) (Englander
and Mayne 2017).
While protein folding is performed several residues are deleted and several resides are added to the original structure
(ribosylation, glycosylation, acylation etc.).
This folding process is carried inside the protein folding complex known as proteasome complex or the chaperone
system.
Both eukaryote and prokaryotic organisms has different types of protein folding complexes.
There are four levels of protein folding: Primary, secondary, tertiary and quarternary structure.
Various types of interactions plays an important role during the protein folding process.
Hydrophobic and Vander walls interactions are the most important interactions playing a significant role in protein
synthesis.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

PART B
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
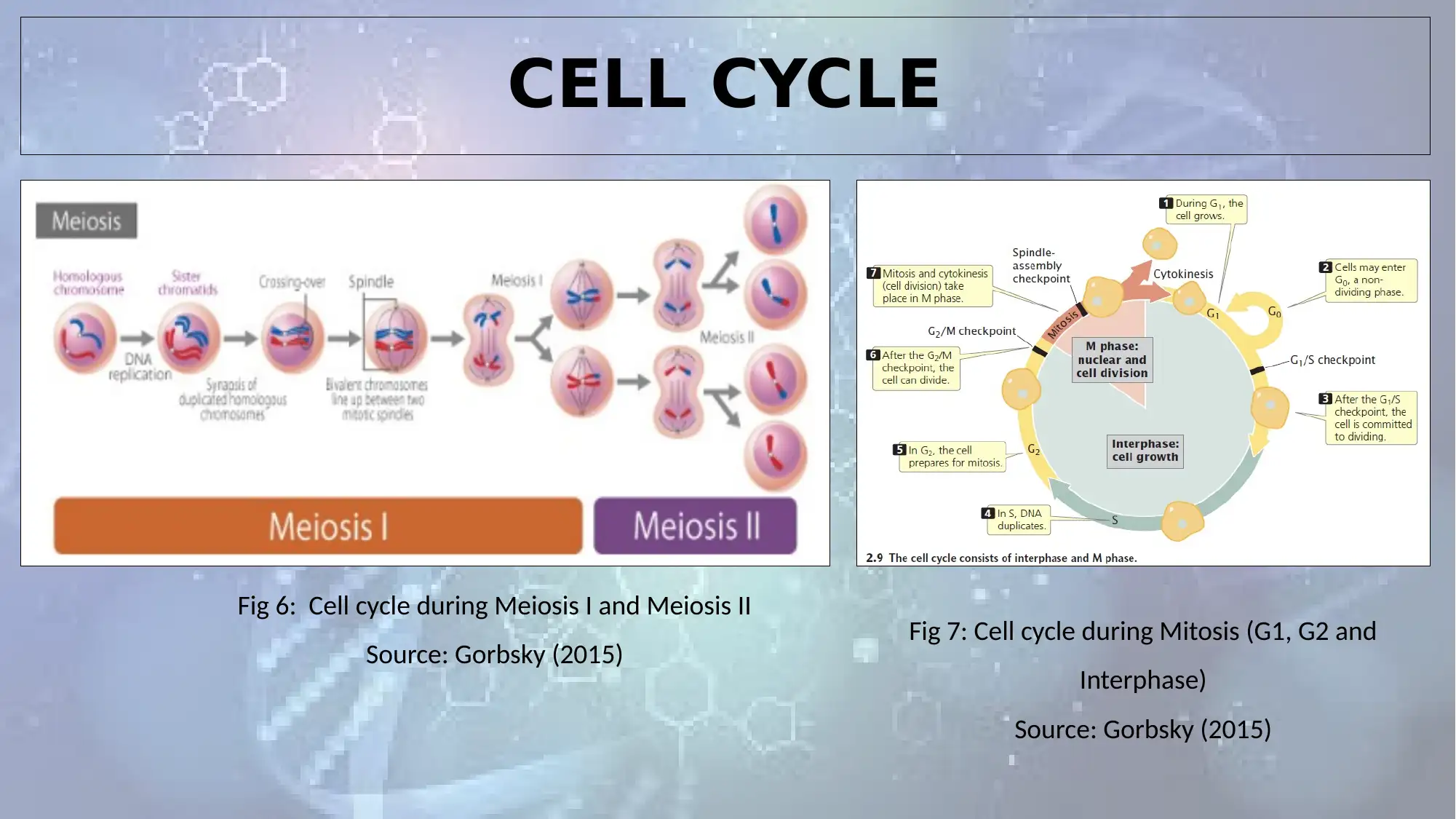
CELL CYCLE
Fig 6: Cell cycle during Meiosis I and Meiosis II
Source: Gorbsky (2015) Fig 7: Cell cycle during Mitosis (G1, G2 and
Interphase)
Source: Gorbsky (2015)
Fig 6: Cell cycle during Meiosis I and Meiosis II
Source: Gorbsky (2015) Fig 7: Cell cycle during Mitosis (G1, G2 and
Interphase)
Source: Gorbsky (2015)
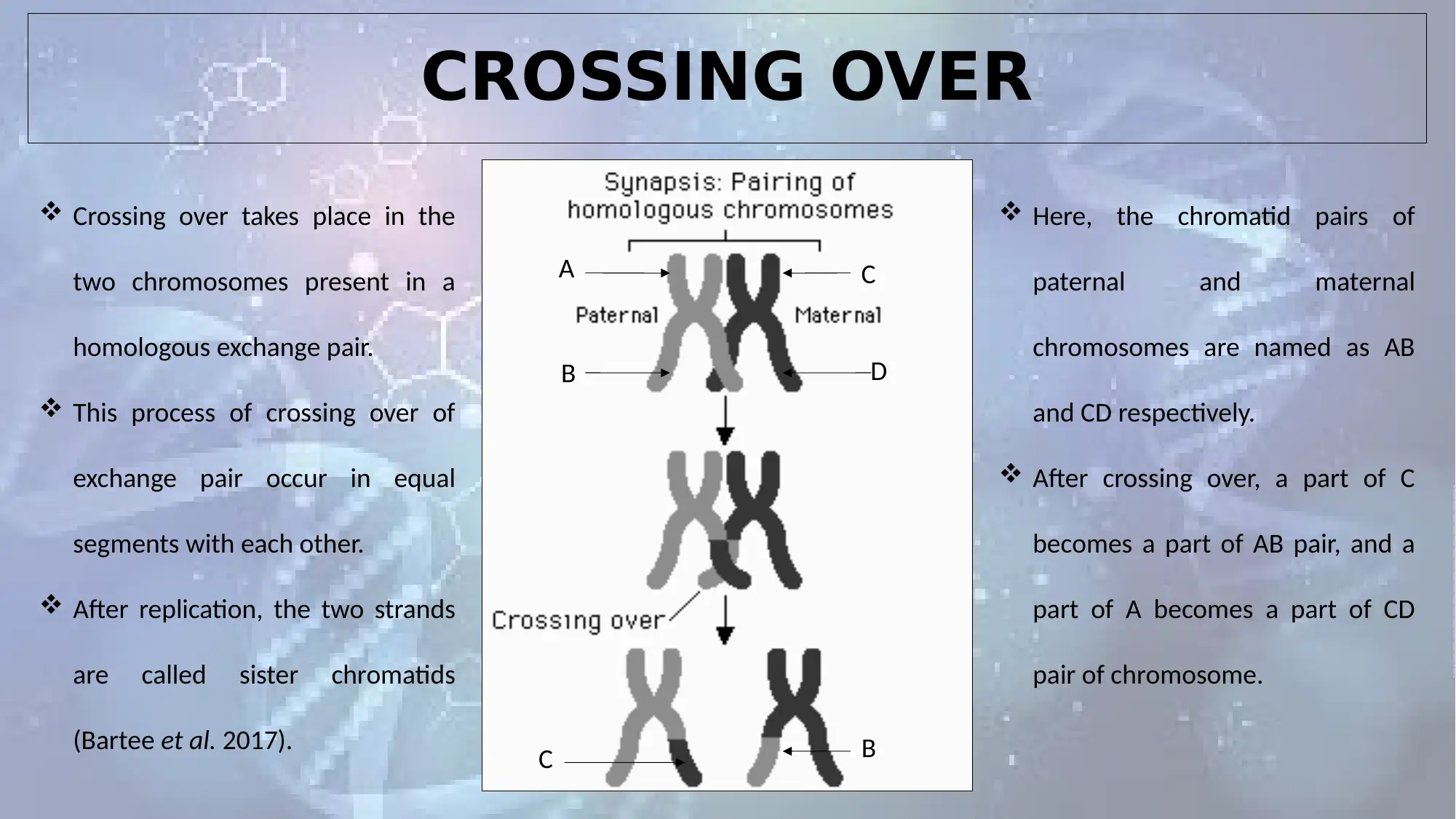
CROSSING OVER
A
B
C
D
BC
Crossing over takes place in the
two chromosomes present in a
homologous exchange pair.
This process of crossing over of
exchange pair occur in equal
segments with each other.
After replication, the two strands
are called sister chromatids
(Bartee et al. 2017).
Here, the chromatid pairs of
paternal and maternal
chromosomes are named as AB
and CD respectively.
After crossing over, a part of C
becomes a part of AB pair, and a
part of A becomes a part of CD
pair of chromosome.
A
B
C
D
BC
Crossing over takes place in the
two chromosomes present in a
homologous exchange pair.
This process of crossing over of
exchange pair occur in equal
segments with each other.
After replication, the two strands
are called sister chromatids
(Bartee et al. 2017).
Here, the chromatid pairs of
paternal and maternal
chromosomes are named as AB
and CD respectively.
After crossing over, a part of C
becomes a part of AB pair, and a
part of A becomes a part of CD
pair of chromosome.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
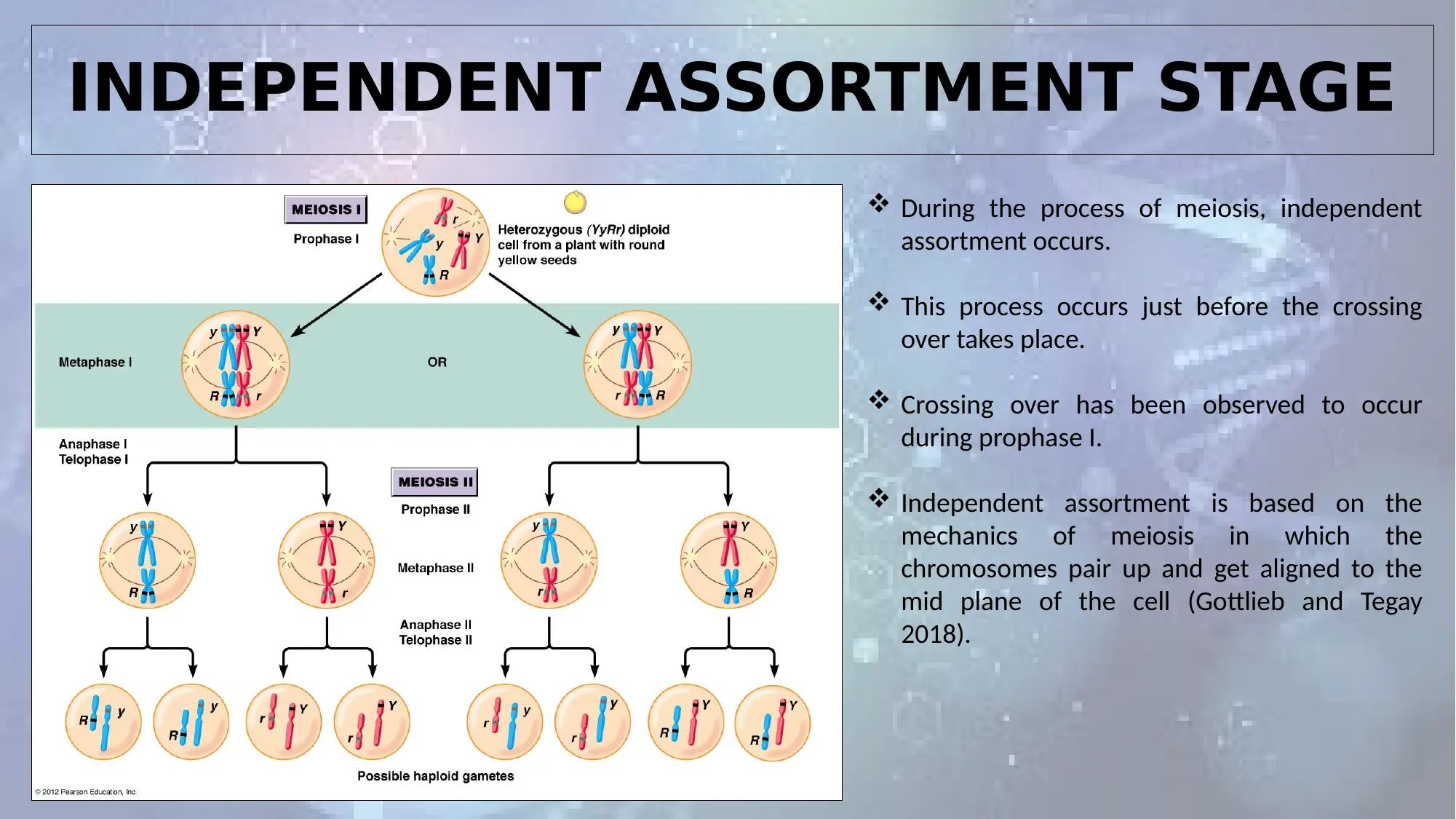
INDEPENDENT ASSORTMENT STAGE
During the process of meiosis, independent
assortment occurs.
This process occurs just before the crossing
over takes place.
Crossing over has been observed to occur
during prophase I.
Independent assortment is based on the
mechanics of meiosis in which the
chromosomes pair up and get aligned to the
mid plane of the cell (Gottlieb and Tegay
2018).
During the process of meiosis, independent
assortment occurs.
This process occurs just before the crossing
over takes place.
Crossing over has been observed to occur
during prophase I.
Independent assortment is based on the
mechanics of meiosis in which the
chromosomes pair up and get aligned to the
mid plane of the cell (Gottlieb and Tegay
2018).
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

RANDOM SEGREGATION
Random segregation occurs when two meiotic chromosomes are segregated or separated randomly into reproductive
gamete cells.
The chromosomal segregation occurs at the two distinguished stages during meiosis (analphase I and anaphase II).
During metaphase I, homologous pairs have been found to place themselves in a random pattern.
In Anaphase I, the centromeres breaks apart, and the homologous chromosomes separates.
The sister chromatids separates during anaphase II.
In telophase, the chromosomes start moving to opposite poles.
Cytokinesis separates the cell into two haploid cells.
Random segregation occurs when two meiotic chromosomes are segregated or separated randomly into reproductive
gamete cells.
The chromosomal segregation occurs at the two distinguished stages during meiosis (analphase I and anaphase II).
During metaphase I, homologous pairs have been found to place themselves in a random pattern.
In Anaphase I, the centromeres breaks apart, and the homologous chromosomes separates.
The sister chromatids separates during anaphase II.
In telophase, the chromosomes start moving to opposite poles.
Cytokinesis separates the cell into two haploid cells.
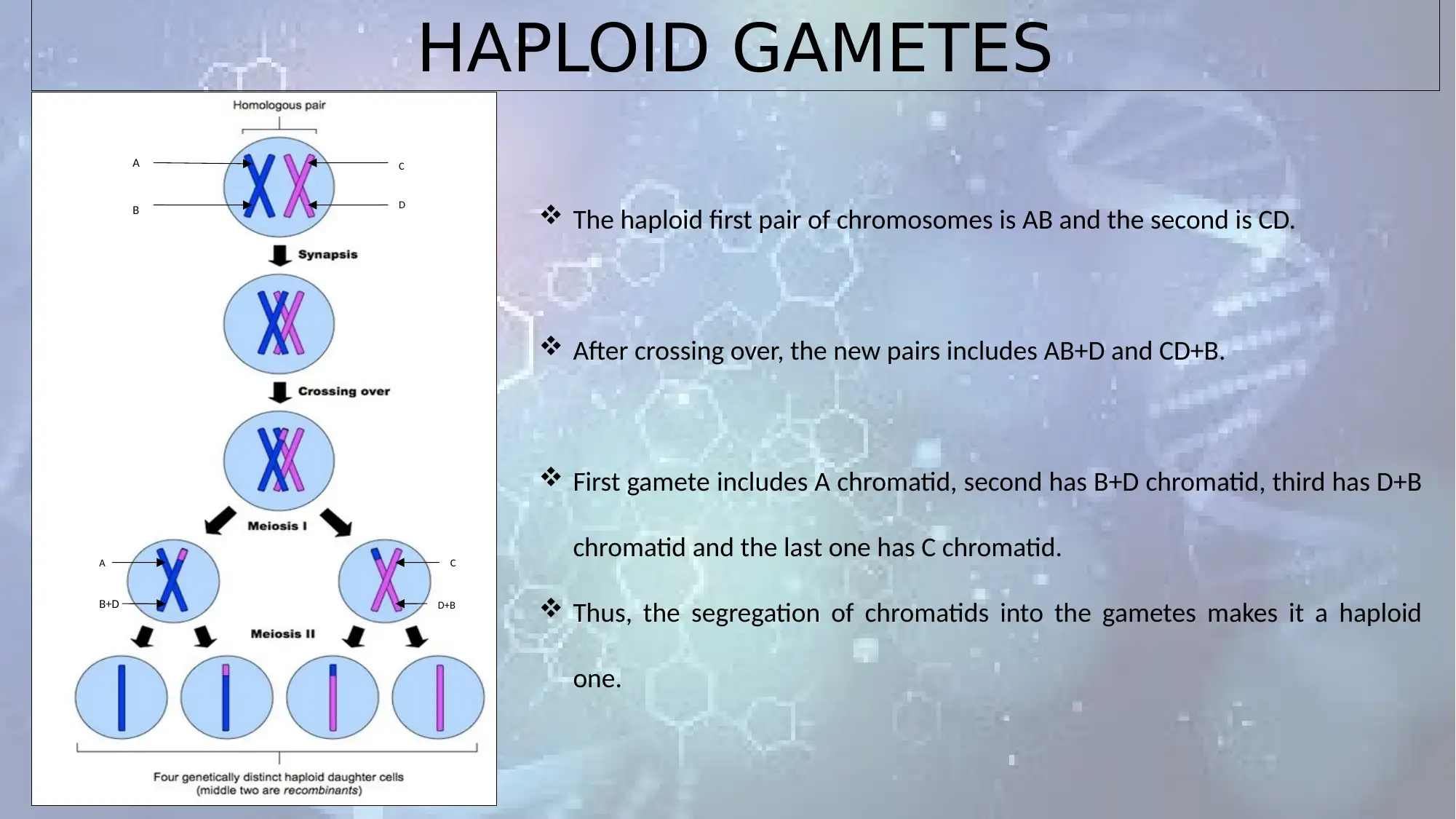
HAPLOID GAMETES
A
B
C
D
A
B+D D+B
C
The haploid first pair of chromosomes is AB and the second is CD.
After crossing over, the new pairs includes AB+D and CD+B.
First gamete includes A chromatid, second has B+D chromatid, third has D+B
chromatid and the last one has C chromatid.
Thus, the segregation of chromatids into the gametes makes it a haploid
one.
A
B
C
D
A
B+D D+B
C
The haploid first pair of chromosomes is AB and the second is CD.
After crossing over, the new pairs includes AB+D and CD+B.
First gamete includes A chromatid, second has B+D chromatid, third has D+B
chromatid and the last one has C chromatid.
Thus, the segregation of chromatids into the gametes makes it a haploid
one.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
1 out of 13
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.




