University of Brighton: BY263 Genomics Assessment on Plasmid DNA
VerifiedAdded on 2022/10/06
|8
|1262
|19
Homework Assignment
AI Summary
This assignment assesses a student's understanding of genomics through an analysis of plasmid DNA. The student performed experiments involving plasmid DNA extraction from bacterial samples, quantification via spectrophotometry, and restriction digestion using the EcoR1 enzyme. The assignment includes calculations of DNA concentration and yield, along with an interpretation of agarose gel electrophoresis results. The student correctly identifies the presence of uncut and digested DNA fragments, including the presence of extra chromosomal DNA in one of the samples. The assignment also includes a scientific abstract summarizing the experimental procedure, results, and conclusions, demonstrating the student's grasp of the techniques and their ability to interpret the data to draw meaningful conclusions about the plasmid DNA and the presence of extra chromosomal DNA.

Running Head: GENOMICS ASSESSMENT
Genomics Assessment
Name of the Student:
Name of the University:
Author note:
Genomics Assessment
Name of the Student:
Name of the University:
Author note:
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
1GENOMICS ASSESSMENT
Task One: Assignment
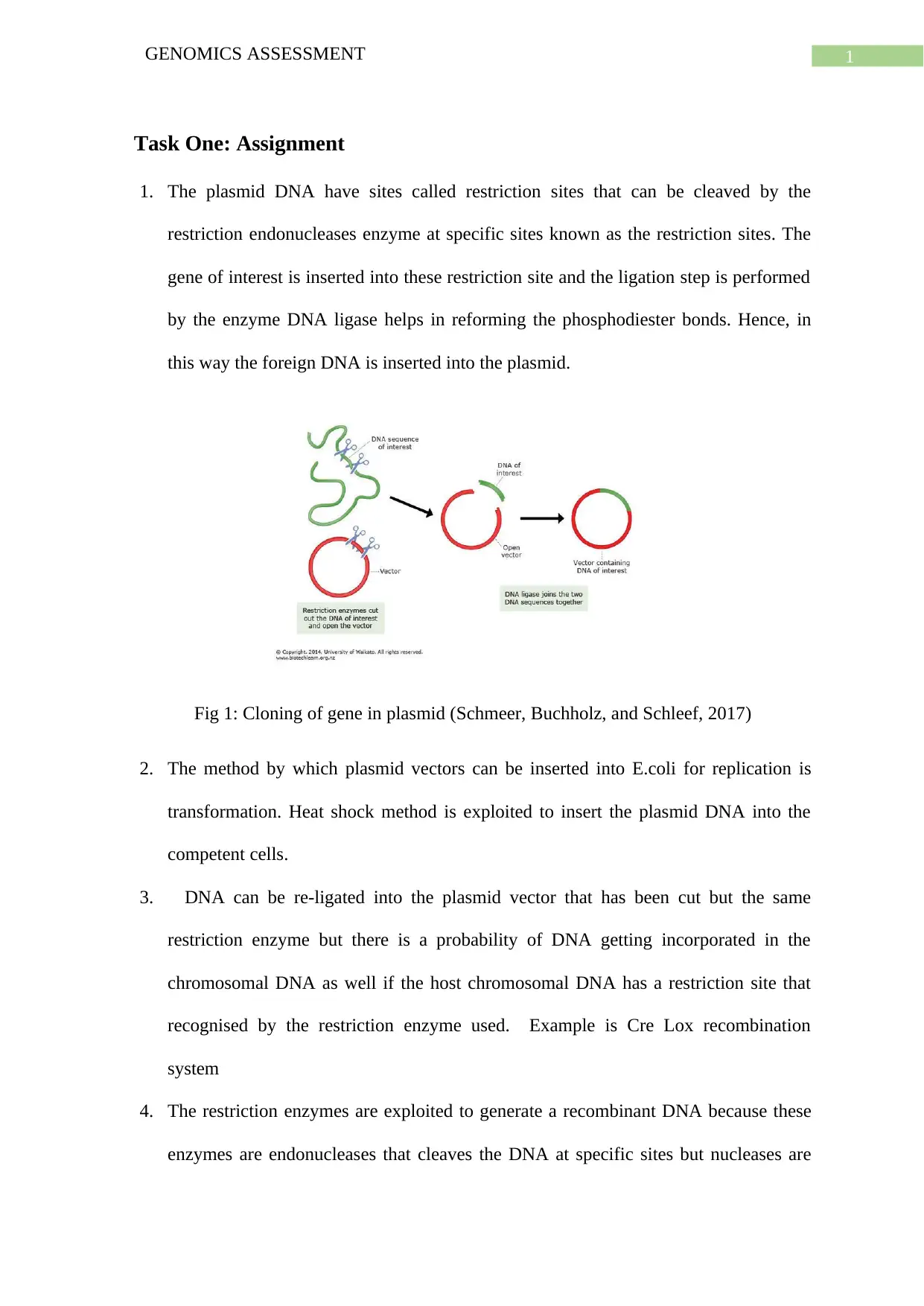
1. The plasmid DNA have sites called restriction sites that can be cleaved by the
restriction endonucleases enzyme at specific sites known as the restriction sites. The
gene of interest is inserted into these restriction site and the ligation step is performed
by the enzyme DNA ligase helps in reforming the phosphodiester bonds. Hence, in
this way the foreign DNA is inserted into the plasmid.
Fig 1: Cloning of gene in plasmid (Schmeer, Buchholz, and Schleef, 2017)
2. The method by which plasmid vectors can be inserted into E.coli for replication is
transformation. Heat shock method is exploited to insert the plasmid DNA into the
competent cells.
3. DNA can be re-ligated into the plasmid vector that has been cut but the same
restriction enzyme but there is a probability of DNA getting incorporated in the
chromosomal DNA as well if the host chromosomal DNA has a restriction site that
recognised by the restriction enzyme used. Example is Cre Lox recombination
system
4. The restriction enzymes are exploited to generate a recombinant DNA because these
enzymes are endonucleases that cleaves the DNA at specific sites but nucleases are
Task One: Assignment
1. The plasmid DNA have sites called restriction sites that can be cleaved by the
restriction endonucleases enzyme at specific sites known as the restriction sites. The
gene of interest is inserted into these restriction site and the ligation step is performed
by the enzyme DNA ligase helps in reforming the phosphodiester bonds. Hence, in
this way the foreign DNA is inserted into the plasmid.
Fig 1: Cloning of gene in plasmid (Schmeer, Buchholz, and Schleef, 2017)
2. The method by which plasmid vectors can be inserted into E.coli for replication is
transformation. Heat shock method is exploited to insert the plasmid DNA into the
competent cells.
3. DNA can be re-ligated into the plasmid vector that has been cut but the same
restriction enzyme but there is a probability of DNA getting incorporated in the
chromosomal DNA as well if the host chromosomal DNA has a restriction site that
recognised by the restriction enzyme used. Example is Cre Lox recombination
system
4. The restriction enzymes are exploited to generate a recombinant DNA because these
enzymes are endonucleases that cleaves the DNA at specific sites but nucleases are

2GENOMICS ASSESSMENT
enzymes that cleave the nucleotides either at the end or in between the strand
(Enghiad and Zhao, 2017).
5.
Sample
name
A260nm A280nm DNA
concentration
(μg/ml)
Yield
(μg)
Ratio
260/280
Plasmid
DNA per cell
C1 0.251 0.097 62.75 3.13 2.58 6.26 x 10-10
C2 0.247 0.083 61.75 3.08 2.98 6.16 x 10-10
The number of cells considered is 5x109 cells
DNA concentration (μg/ml) of C1 = A260 x 50 x dilution factor
= 0.251 x 50 x 5
= 62.75 μg/ml
DNA concentration (μg/ml) of C2 = A260 x 50 x dilution factor
= 0.247 x 50 x 5
= 61.75 μg/ml
DNA yield of C1 = Concentration of DNA x Final volume of the sample
= 62.75 x 0.05
= 3.13 μg
DNA yield of C2 = Concentration of DNA x Final volume of the sample
= 61.75 x 0.05
= 3.08 μg
The amount of plasmid DNA per cell is = (3.13/5 x 109)
= 6.26 x 10-10
The amount of plasmid DNA per cell is = (3.08/5 x 109)
enzymes that cleave the nucleotides either at the end or in between the strand
(Enghiad and Zhao, 2017).
5.
Sample
name
A260nm A280nm DNA
concentration
(μg/ml)
Yield
(μg)
Ratio
260/280
Plasmid
DNA per cell
C1 0.251 0.097 62.75 3.13 2.58 6.26 x 10-10
C2 0.247 0.083 61.75 3.08 2.98 6.16 x 10-10
The number of cells considered is 5x109 cells
DNA concentration (μg/ml) of C1 = A260 x 50 x dilution factor
= 0.251 x 50 x 5
= 62.75 μg/ml
DNA concentration (μg/ml) of C2 = A260 x 50 x dilution factor
= 0.247 x 50 x 5
= 61.75 μg/ml
DNA yield of C1 = Concentration of DNA x Final volume of the sample
= 62.75 x 0.05
= 3.13 μg
DNA yield of C2 = Concentration of DNA x Final volume of the sample
= 61.75 x 0.05
= 3.08 μg
The amount of plasmid DNA per cell is = (3.13/5 x 109)
= 6.26 x 10-10
The amount of plasmid DNA per cell is = (3.08/5 x 109)
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide
3GENOMICS ASSESSMENT
= 6.16 x 10-10
6. 1:10 dilution mean 1 part out of 10 parts.
The final volume given is 50 μL
50 μL = 0.05mL since 1mL= 1000 μL
Therefore, 1:10 of 0.05mL = 0.005mL
0.005mL = 5 μL
Answer: 1:10 dilution of 50 μL means 45 μL of water and 5 μL of the chemical.
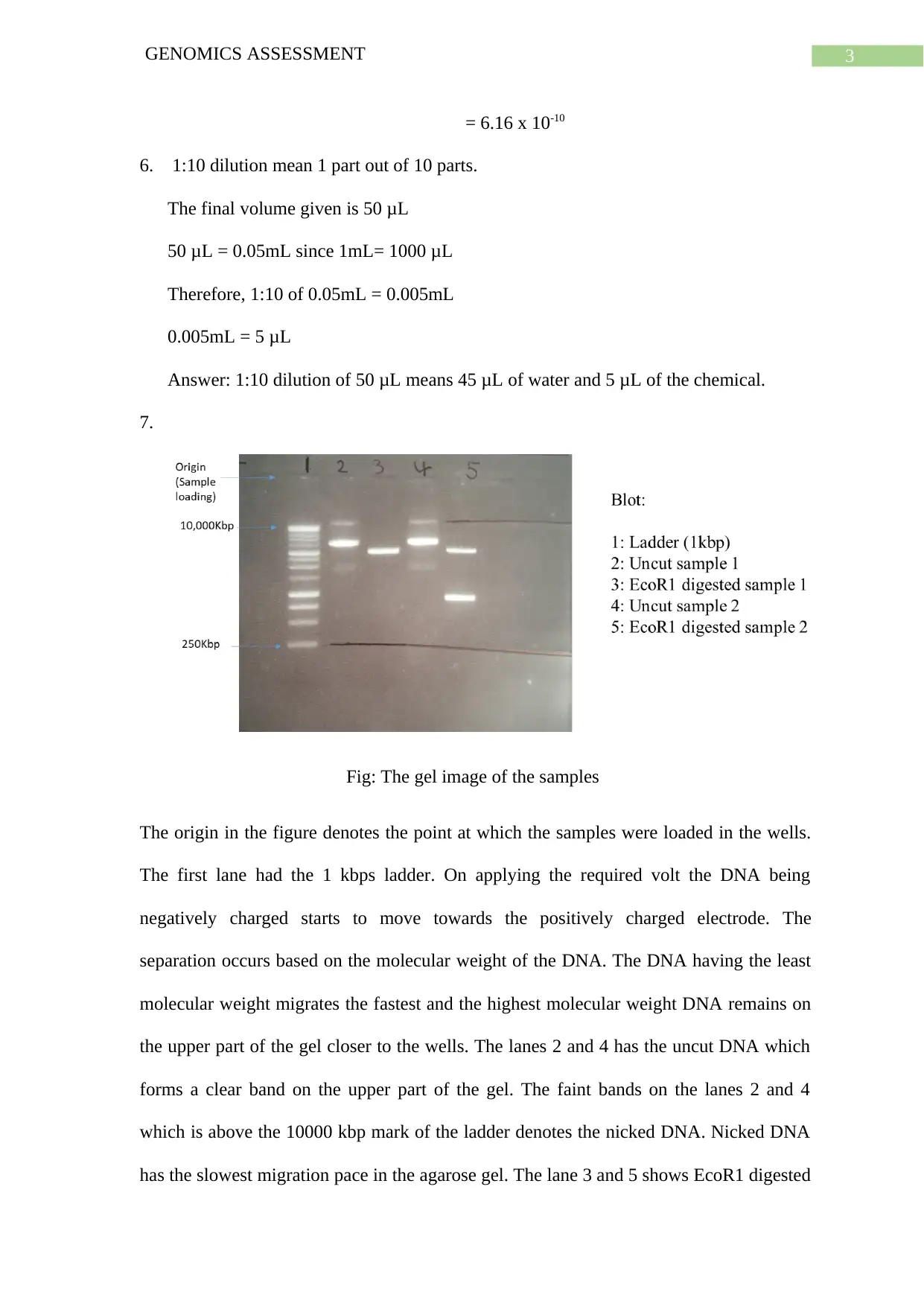
7.
Fig: The gel image of the samples
The origin in the figure denotes the point at which the samples were loaded in the wells.
The first lane had the 1 kbps ladder. On applying the required volt the DNA being
negatively charged starts to move towards the positively charged electrode. The
separation occurs based on the molecular weight of the DNA. The DNA having the least
molecular weight migrates the fastest and the highest molecular weight DNA remains on
the upper part of the gel closer to the wells. The lanes 2 and 4 has the uncut DNA which
forms a clear band on the upper part of the gel. The faint bands on the lanes 2 and 4
which is above the 10000 kbp mark of the ladder denotes the nicked DNA. Nicked DNA
has the slowest migration pace in the agarose gel. The lane 3 and 5 shows EcoR1 digested
= 6.16 x 10-10
6. 1:10 dilution mean 1 part out of 10 parts.
The final volume given is 50 μL
50 μL = 0.05mL since 1mL= 1000 μL
Therefore, 1:10 of 0.05mL = 0.005mL
0.005mL = 5 μL
Answer: 1:10 dilution of 50 μL means 45 μL of water and 5 μL of the chemical.
7.
Fig: The gel image of the samples
The origin in the figure denotes the point at which the samples were loaded in the wells.
The first lane had the 1 kbps ladder. On applying the required volt the DNA being
negatively charged starts to move towards the positively charged electrode. The
separation occurs based on the molecular weight of the DNA. The DNA having the least
molecular weight migrates the fastest and the highest molecular weight DNA remains on
the upper part of the gel closer to the wells. The lanes 2 and 4 has the uncut DNA which
forms a clear band on the upper part of the gel. The faint bands on the lanes 2 and 4
which is above the 10000 kbp mark of the ladder denotes the nicked DNA. Nicked DNA
has the slowest migration pace in the agarose gel. The lane 3 and 5 shows EcoR1 digested
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

4GENOMICS ASSESSMENT
sample 1 and 2. The sample 1 had one cut site so a single intense band is obtained. But
sample 2 has 2 cut sites and hence 2 distinct bands are obtained in the lane 5. The bigger
fragment had lesser mobility and hence is on the upper part of the gel and the smaller
band has migrated more.
8. Both the sample 1 and sample 2 contains plasmid. And plasmid in sample 2 contains
extra chromosomal DNA. The size of the plasmid DNA is approximately between
5000 to 4000 Kbp. The size of the extra chromosomal DNA is approximately 1000
kbp.
Task Two: Scientific Abstract
Restriction digestion is a process exploited to prepare a DNA for various analytical
purposes. The process cleaves the DNA at specific sites known as the restriction site with the
help of specific enzymes called restriction enzymes (Green and Sambrook, 2016). This
experiment mainly focuses on the restriction digestion of the selected and prepared plasmid
DNA. The main objective is to identify the presence or absence of newly emerged pathogenic
bacterial strains.
The method exploited to execute the experiment involved preparation of the plasmid
DNA. There were two bacterial strains considered from two samples C1 and C2. The cultures
ere pelleted and passed on for plasmid extraction, quantification and identification. Miniprep
method was used to extract DNA from bacterial cells. The yield and purity of the extracted
DNA was determined by spectrophotometric methods. The restriction digestion of the
purified DNA was carried out using EcoR1 restriction enzyme. Agarose Gel Electrophoresis
of the digested DNA was carried out keeping the uncut DNA as the control.
The DNA obtained from both the samples C1 and C2 had a high purity of 2.58 and
2.98 respectively. Based on the UV absorbance data obtained at 260nm and 280nm these data
sample 1 and 2. The sample 1 had one cut site so a single intense band is obtained. But
sample 2 has 2 cut sites and hence 2 distinct bands are obtained in the lane 5. The bigger
fragment had lesser mobility and hence is on the upper part of the gel and the smaller
band has migrated more.
8. Both the sample 1 and sample 2 contains plasmid. And plasmid in sample 2 contains
extra chromosomal DNA. The size of the plasmid DNA is approximately between
5000 to 4000 Kbp. The size of the extra chromosomal DNA is approximately 1000
kbp.
Task Two: Scientific Abstract
Restriction digestion is a process exploited to prepare a DNA for various analytical
purposes. The process cleaves the DNA at specific sites known as the restriction site with the
help of specific enzymes called restriction enzymes (Green and Sambrook, 2016). This
experiment mainly focuses on the restriction digestion of the selected and prepared plasmid
DNA. The main objective is to identify the presence or absence of newly emerged pathogenic
bacterial strains.
The method exploited to execute the experiment involved preparation of the plasmid
DNA. There were two bacterial strains considered from two samples C1 and C2. The cultures
ere pelleted and passed on for plasmid extraction, quantification and identification. Miniprep
method was used to extract DNA from bacterial cells. The yield and purity of the extracted
DNA was determined by spectrophotometric methods. The restriction digestion of the
purified DNA was carried out using EcoR1 restriction enzyme. Agarose Gel Electrophoresis
of the digested DNA was carried out keeping the uncut DNA as the control.
The DNA obtained from both the samples C1 and C2 had a high purity of 2.58 and
2.98 respectively. Based on the UV absorbance data obtained at 260nm and 280nm these data

5GENOMICS ASSESSMENT
was obtained (Green and Sambrook, 2018). The quantitation revealed from the absorbance
data is that approximately 6 x 10-10 plasmid DNA per cell was obtained. This extracted DNA
was taken for restriction digestion post which it was run in agarose gel. The restriction
enzyme used was EcoR1. The gel image showed the presence of a clear band of the sample 1
after cleaving with the restriction enzyme. The second sample had 2 clear bands after
restriction digestion indicating that there were two cuts. The control samples which were the
untreated samples had two bands for the intact DNA having low migration rate owing to the
high molecular weight. The faint bands were also observed which were due to presence of the
nicked DNA.
It can be concluded from this experiment that the DNA pure extracted DNA that was
digested yielded the desired result in sample 1 but sample 2 had two cuts indicating the
presence of extra chromosomal DNA.
was obtained (Green and Sambrook, 2018). The quantitation revealed from the absorbance
data is that approximately 6 x 10-10 plasmid DNA per cell was obtained. This extracted DNA
was taken for restriction digestion post which it was run in agarose gel. The restriction
enzyme used was EcoR1. The gel image showed the presence of a clear band of the sample 1
after cleaving with the restriction enzyme. The second sample had 2 clear bands after
restriction digestion indicating that there were two cuts. The control samples which were the
untreated samples had two bands for the intact DNA having low migration rate owing to the
high molecular weight. The faint bands were also observed which were due to presence of the
nicked DNA.
It can be concluded from this experiment that the DNA pure extracted DNA that was
digested yielded the desired result in sample 1 but sample 2 had two cuts indicating the
presence of extra chromosomal DNA.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

6GENOMICS ASSESSMENT
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

7GENOMICS ASSESSMENT
References
Green, M. R., and Sambrook, J., 2016. Preparation of plasmid DNA by alkaline lysis with
sodium dodecyl sulfate: minipreps. Cold Spring Harbor Protocols, 2016(10), pdb-
prot093344.
Enghiad, B., and Zhao, H., 2017. Programmable DNA-guided artificial restriction
enzymes. ACS synthetic biology, 6(5), 752-757.
Schmeer, M., Buchholz, T., and Schleef, M., 2017. Plasmid DNA manufacturing for indirect
and direct clinical applications. Human gene therapy, 28(10), 856-861.
Green, M.R. and Sambrook, J., 2018. Isolation and quantification of DNA. Cold Spring
Harbor Protocols, 2018(6), pp.pdb-top093336.
References
Green, M. R., and Sambrook, J., 2016. Preparation of plasmid DNA by alkaline lysis with
sodium dodecyl sulfate: minipreps. Cold Spring Harbor Protocols, 2016(10), pdb-
prot093344.
Enghiad, B., and Zhao, H., 2017. Programmable DNA-guided artificial restriction
enzymes. ACS synthetic biology, 6(5), 752-757.
Schmeer, M., Buchholz, T., and Schleef, M., 2017. Plasmid DNA manufacturing for indirect
and direct clinical applications. Human gene therapy, 28(10), 856-861.
Green, M.R. and Sambrook, J., 2018. Isolation and quantification of DNA. Cold Spring
Harbor Protocols, 2018(6), pp.pdb-top093336.
1 out of 8
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.