Structure, Function, and Analysis of Proteins: Enzyme Kinetics Report
VerifiedAdded on 2021/08/11
|8
|2187
|316
Report
AI Summary
This report details an experiment on the enzyme kinetics of Yeast Alcohol Dehydrogenase (YAD). The study investigates the enzyme's activity with various substrates, including ethanol, propan-1-ol, and butan-1-ol. The methodology involves standard assay conditions, spectrophotometric measurements, and Lineweaver-Burk plot analysis. The results reveal that some alcohols act as substrates while others, like 2-chloroethanol, act as competitive inhibitors. The report discusses the impact of substrate structure, hydrophobicity, and steric factors on enzyme activity and binding affinity. It also examines the effect of a competitive inhibitor on the enzyme's kinetic parameters, including Km and Vmax. The findings highlight the enzyme's specificity and the mechanisms of alcohol oxidation and inhibition.

Structure, Function and Analysis of
Proteins (LIFE2087)
Title: Experiment on enzyme kinetics of Yeast
Alcohol Dehydrogenase and its substrate
properties
Name: Priangka Ambheghai Bathianathan
Student ID: 20210653
Proteins (LIFE2087)
Title: Experiment on enzyme kinetics of Yeast
Alcohol Dehydrogenase and its substrate
properties
Name: Priangka Ambheghai Bathianathan
Student ID: 20210653
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Page 2 of 8
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
ABSTRACT
Yeast Alcohol Dehydrogenase (YAD) acts as a catalyst in the oxidation of alcohol. The aim of
this experiment is to study the enzyme kinetics of YAD and how different substrates affect
enzyme activity. Methodology uses standard assay conditions for steady state reaction and
spectrophotometer to measure reaction progress. It was found that out of the 9 substrates
used, Propan-1-ol, Butan-1-ol, Propan-2-ol, 2-Propen-1-ol and 1-Cyclopropyl-methanol act as
substrates whereas Methanol, 2-Chloroethanol, and 1,2-Ethanediol act as non-substrate with
2-chloroethanol also being a competitive inhibitor to the enzyme.
INTRODUCTION
Yeast Alcohol Dehydrogenase (YAD) is a tetramer that works as a catalyst to facilitate
conversion of alcohol to aldehydes or ketones with reduction of Nicotinamide adenine
dinucleotide (NAD+) to NADH.
The main reaction of alcohol dehydrogenase is whereby an alcohol group is oxidized
by the removal of a proton from the hydroxyl group with the help of zinc ions. This is done via
the transfer of a hydride ion from the adjacent carbon atom to NAD+. This NAD+ has an
absorbance of 340nm in a spectrophotometer which is used to study the steady state kinetics
of yeast alcohol dehydrogenase (YAD) in this experiment. The reaction is shown below:
R-CH2OH + NAD+ <----> R-CHO + NADH + H+
This experiment aims to study YAD enzyme and its substrate by 4 parts. First, the initial
rate of enzyme reaction is measured under steady state conditions. Second, the enzyme
activity is measured with different alcohols. Thirdly, the alcohols are studied to determine if
they are substrates of the enzyme or not. Fourthly, the non substrates are further analyzed
to determine if they also act as an inhibitor to the YAD enzyme.
METHOD
This experiment is divided into 4 parts. The first part, Part A is to determine initial rate of YAD
enzyme with ethanol as a substrate under standard assay conditions. The standard assay
condition uses 2.2ml H20, 10mmol/L Pyrophosphate buffer at pH 8.5, 1mmol/L NAD, ethanol
at final concentration of 200mmol/L and YAD at 5.7x10-4mg/ml. The final volume for the assay
is fixed at 3ml for all parts. Absorbance reading is taken, and the initial rate can be calculated.
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
ABSTRACT
Yeast Alcohol Dehydrogenase (YAD) acts as a catalyst in the oxidation of alcohol. The aim of
this experiment is to study the enzyme kinetics of YAD and how different substrates affect
enzyme activity. Methodology uses standard assay conditions for steady state reaction and
spectrophotometer to measure reaction progress. It was found that out of the 9 substrates
used, Propan-1-ol, Butan-1-ol, Propan-2-ol, 2-Propen-1-ol and 1-Cyclopropyl-methanol act as
substrates whereas Methanol, 2-Chloroethanol, and 1,2-Ethanediol act as non-substrate with
2-chloroethanol also being a competitive inhibitor to the enzyme.
INTRODUCTION
Yeast Alcohol Dehydrogenase (YAD) is a tetramer that works as a catalyst to facilitate
conversion of alcohol to aldehydes or ketones with reduction of Nicotinamide adenine
dinucleotide (NAD+) to NADH.
The main reaction of alcohol dehydrogenase is whereby an alcohol group is oxidized
by the removal of a proton from the hydroxyl group with the help of zinc ions. This is done via
the transfer of a hydride ion from the adjacent carbon atom to NAD+. This NAD+ has an
absorbance of 340nm in a spectrophotometer which is used to study the steady state kinetics
of yeast alcohol dehydrogenase (YAD) in this experiment. The reaction is shown below:
R-CH2OH + NAD+ <----> R-CHO + NADH + H+
This experiment aims to study YAD enzyme and its substrate by 4 parts. First, the initial
rate of enzyme reaction is measured under steady state conditions. Second, the enzyme
activity is measured with different alcohols. Thirdly, the alcohols are studied to determine if
they are substrates of the enzyme or not. Fourthly, the non substrates are further analyzed
to determine if they also act as an inhibitor to the YAD enzyme.
METHOD
This experiment is divided into 4 parts. The first part, Part A is to determine initial rate of YAD
enzyme with ethanol as a substrate under standard assay conditions. The standard assay
condition uses 2.2ml H20, 10mmol/L Pyrophosphate buffer at pH 8.5, 1mmol/L NAD, ethanol
at final concentration of 200mmol/L and YAD at 5.7x10-4mg/ml. The final volume for the assay
is fixed at 3ml for all parts. Absorbance reading is taken, and the initial rate can be calculated.
Page 3 of 8
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
Part B is to investigate different alcohols’ ability to act as a substrate for the enzyme
using 9 types of different alcohols. The basic standard assay condition is used but instead of
ethanol at 200mmol/L, each of the different alcohols are used in combination of 0.1ml of
ethanol. Final concentration of the alcohol mixture is still fixed at 200mmol/L. The absorbance
reading is taken to identify which alcohol is a substrate and which is a non-substrate.
Part C is specific to only the non-substrate alcohols identified from part B. This part is
done to study if the non-substrate alcohol also acts as an enzyme inhibitor by comparing
assays with addition of non-substrate and assay of just ethanol. To do this, basic standard
assay is used but 200mmol/L non substrate alcohol is added, and final ethanol concentration
is changed to 10mmol/L. For the only ethanol assay, the 200mmol/L non substrate is changed
to water instead. The absorbance reading is taken to study which non-substrate is also an
inhibitor.
Part D is to determine enzyme kinetic parameters. This is done using the basic
standard assay, but 1mmol/L inhibitor is added, and using 5 different ethanol concentrations.
The absorbance reading is taken, and data is analysed with Lineweaver-Burk plot
RESULTS AND DISCUSSION
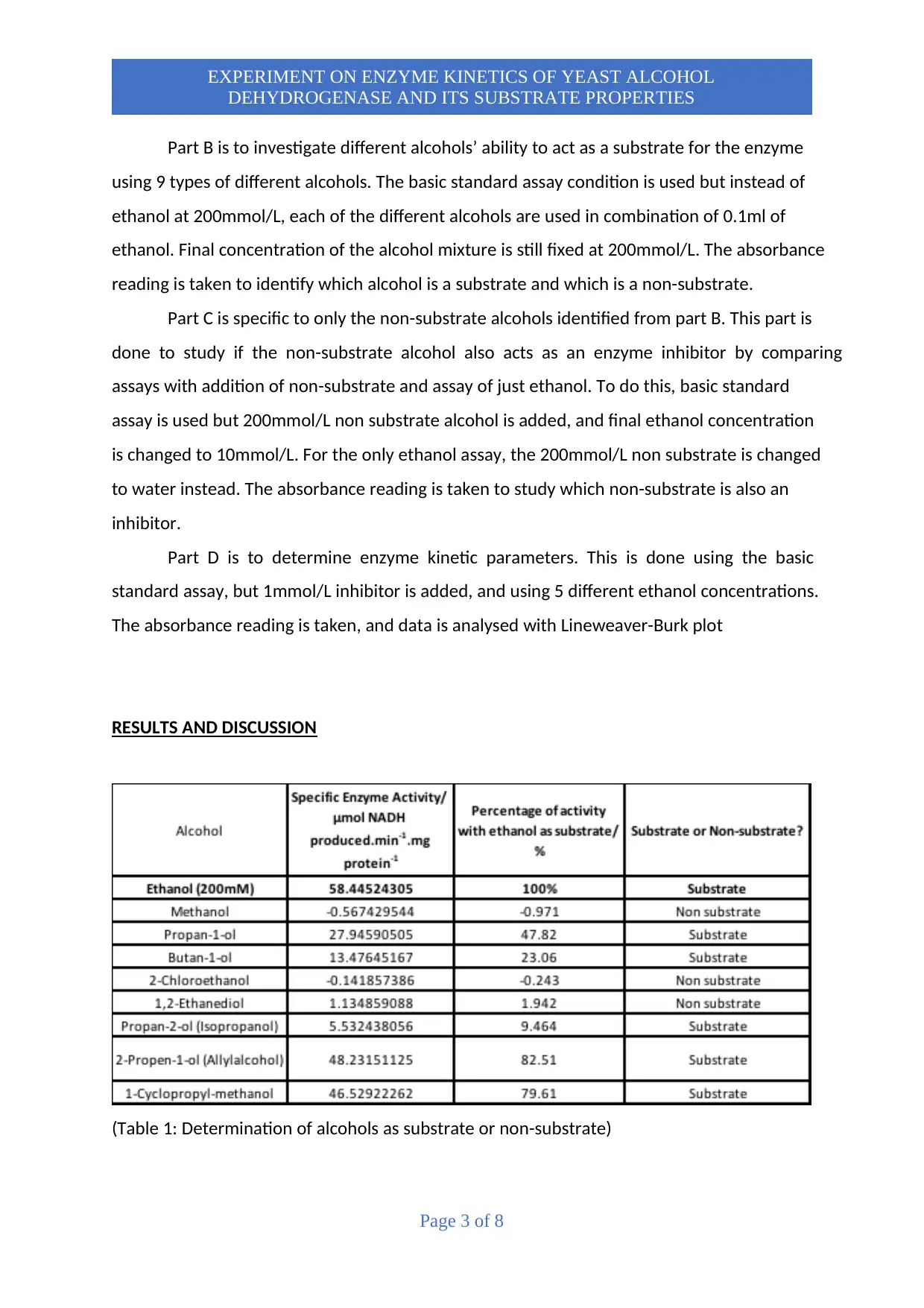
(Table 1: Determination of alcohols as substrate or non-substrate)
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
Part B is to investigate different alcohols’ ability to act as a substrate for the enzyme
using 9 types of different alcohols. The basic standard assay condition is used but instead of
ethanol at 200mmol/L, each of the different alcohols are used in combination of 0.1ml of
ethanol. Final concentration of the alcohol mixture is still fixed at 200mmol/L. The absorbance
reading is taken to identify which alcohol is a substrate and which is a non-substrate.
Part C is specific to only the non-substrate alcohols identified from part B. This part is
done to study if the non-substrate alcohol also acts as an enzyme inhibitor by comparing
assays with addition of non-substrate and assay of just ethanol. To do this, basic standard
assay is used but 200mmol/L non substrate alcohol is added, and final ethanol concentration
is changed to 10mmol/L. For the only ethanol assay, the 200mmol/L non substrate is changed
to water instead. The absorbance reading is taken to study which non-substrate is also an
inhibitor.
Part D is to determine enzyme kinetic parameters. This is done using the basic
standard assay, but 1mmol/L inhibitor is added, and using 5 different ethanol concentrations.
The absorbance reading is taken, and data is analysed with Lineweaver-Burk plot
RESULTS AND DISCUSSION
(Table 1: Determination of alcohols as substrate or non-substrate)
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Page 4 of 8
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
YAD enzymes consists of predominant ADH1 which works to oxidize aliphatic unbranched
alcohols. This explains why the unbranched aliphatic alcohols from table 1 such as Ethanol,
Propan-1-ol, Butan-1-ol, and 2-propen-1-ol are good substrates. It has been studied that
activity of Yeast Alcohol Dehydrogenase decreases with increasing chain length due to
increased hydrophobicity. Thus, this explains why among the unbranched aliphatic alcohols,
Ethanol shows highest enzyme activity due to it being the shortest alcohol chain.
In addition, it also shows that YAD works less effectively moving from primary to
secondary alcohol which is further validated in the experiment whereby enzyme activity is
higher in Propan-1-ol compared to Propan-2-ol due to Propan-2-ol being a secondary alcohol
which weakens the binding of NAD+. This can be because the oxidation of secondary alcohols
causes destruction of chirality while it transforms sp3-hybridized alcohols into sp2 hybridized
carbonyl groups.
To understand why 2-Propen-1-ol is a good substrate with a high enzyme activity, we
must understand the structure of the substrate. This substrate is an allyl alcohol. It is a small
unsaturated alcohol thus it has higher hydrophilicity and lower hydrophobicity which allows
more effective binding.
1-cyclopropyl-methanol is also a good substrate for YAD. To explain this, we must first
understand the structure of the enzyme. The subunits of the enzymes are divided into
catalytic domain and coenzyme-binding domain. The domains are separated by a cleft which
has a deep pocket known as the substrate binding pocket which consists of the substrate and
the coenzyme. Thus, it has been studied that an alcohol substrate with a bulkier substituent
such as a cyclopropane will cause more effective binding due to better fitting.
The major effect of the substrate specificity is based on substituents. Electron
withdrawing substituents such as halogens slow down alcohol oxidation. This is because the
environment in nicotinamide ring of the enzyme is positively charged which is essential for
effective binding. This explains 2-chloroethanol being a non-substrate due to the chlorine
atom.
It shows that methanol as a substrate for alcohol dehydrogenase indicated a very slow
rate of reaction. Due to the lack of hydrophobic chain, the binding of the substrate to the
enzyme is unfavored because in the enzyme-coenzyme substrate complex, the methyl group
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
YAD enzymes consists of predominant ADH1 which works to oxidize aliphatic unbranched
alcohols. This explains why the unbranched aliphatic alcohols from table 1 such as Ethanol,
Propan-1-ol, Butan-1-ol, and 2-propen-1-ol are good substrates. It has been studied that
activity of Yeast Alcohol Dehydrogenase decreases with increasing chain length due to
increased hydrophobicity. Thus, this explains why among the unbranched aliphatic alcohols,
Ethanol shows highest enzyme activity due to it being the shortest alcohol chain.
In addition, it also shows that YAD works less effectively moving from primary to
secondary alcohol which is further validated in the experiment whereby enzyme activity is
higher in Propan-1-ol compared to Propan-2-ol due to Propan-2-ol being a secondary alcohol
which weakens the binding of NAD+. This can be because the oxidation of secondary alcohols
causes destruction of chirality while it transforms sp3-hybridized alcohols into sp2 hybridized
carbonyl groups.
To understand why 2-Propen-1-ol is a good substrate with a high enzyme activity, we
must understand the structure of the substrate. This substrate is an allyl alcohol. It is a small
unsaturated alcohol thus it has higher hydrophilicity and lower hydrophobicity which allows
more effective binding.
1-cyclopropyl-methanol is also a good substrate for YAD. To explain this, we must first
understand the structure of the enzyme. The subunits of the enzymes are divided into
catalytic domain and coenzyme-binding domain. The domains are separated by a cleft which
has a deep pocket known as the substrate binding pocket which consists of the substrate and
the coenzyme. Thus, it has been studied that an alcohol substrate with a bulkier substituent
such as a cyclopropane will cause more effective binding due to better fitting.
The major effect of the substrate specificity is based on substituents. Electron
withdrawing substituents such as halogens slow down alcohol oxidation. This is because the
environment in nicotinamide ring of the enzyme is positively charged which is essential for
effective binding. This explains 2-chloroethanol being a non-substrate due to the chlorine
atom.
It shows that methanol as a substrate for alcohol dehydrogenase indicated a very slow
rate of reaction. Due to the lack of hydrophobic chain, the binding of the substrate to the
enzyme is unfavored because in the enzyme-coenzyme substrate complex, the methyl group
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser
Page 5 of 8
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
substrate is too small and cannot be held in a fixed position which causes low hydride transfer
rate.
Finally, 1,2-Ethanediol is not a good substrate because diols are optically active and
sterically demanding. Thus, YAD enzyme has its own regioselectivity and is unable to
effectively bind many of the substrate molecules due to the steric demand.
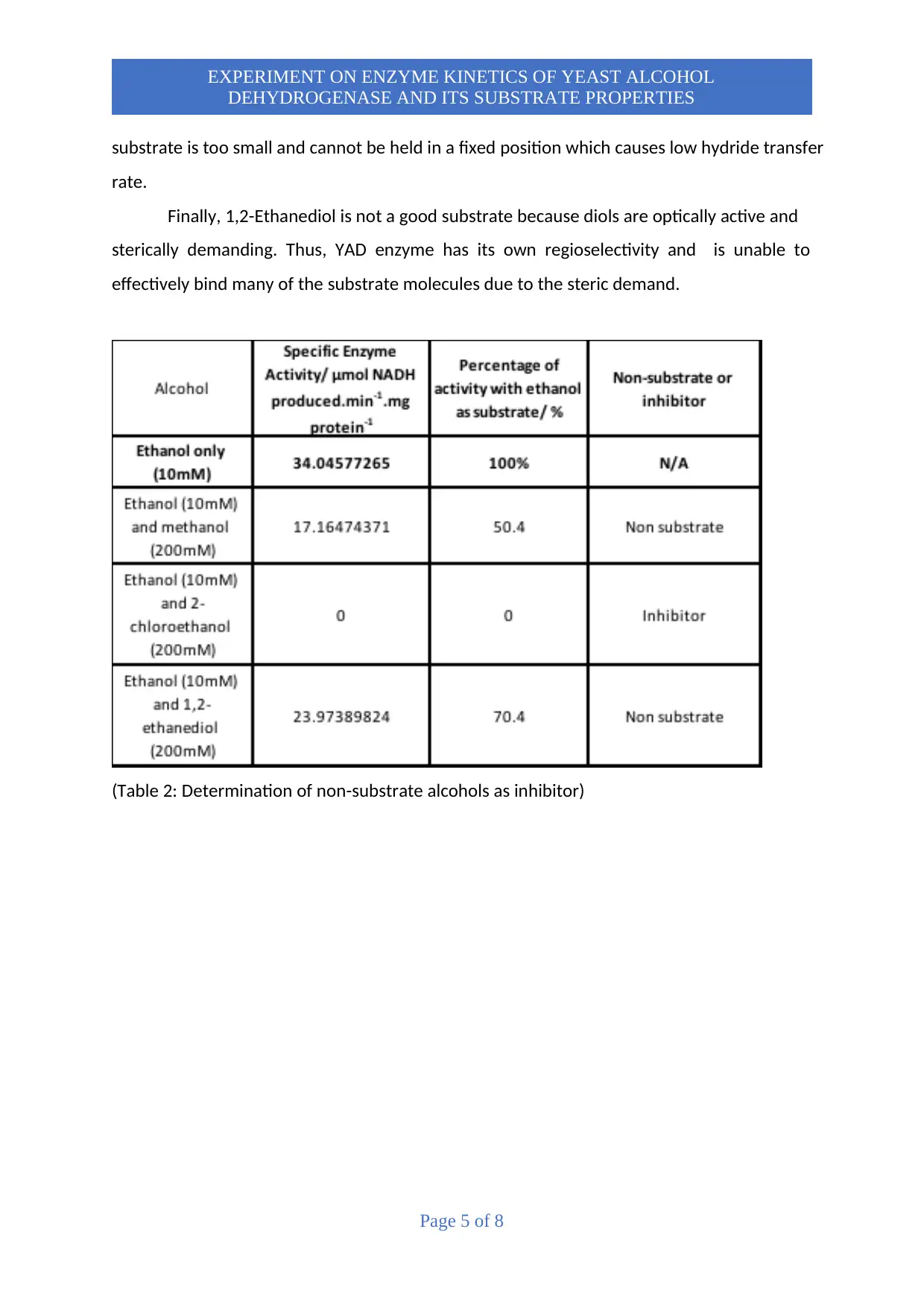
(Table 2: Determination of non-substrate alcohols as inhibitor)
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
substrate is too small and cannot be held in a fixed position which causes low hydride transfer
rate.
Finally, 1,2-Ethanediol is not a good substrate because diols are optically active and
sterically demanding. Thus, YAD enzyme has its own regioselectivity and is unable to
effectively bind many of the substrate molecules due to the steric demand.
(Table 2: Determination of non-substrate alcohols as inhibitor)
Page 6 of 8
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
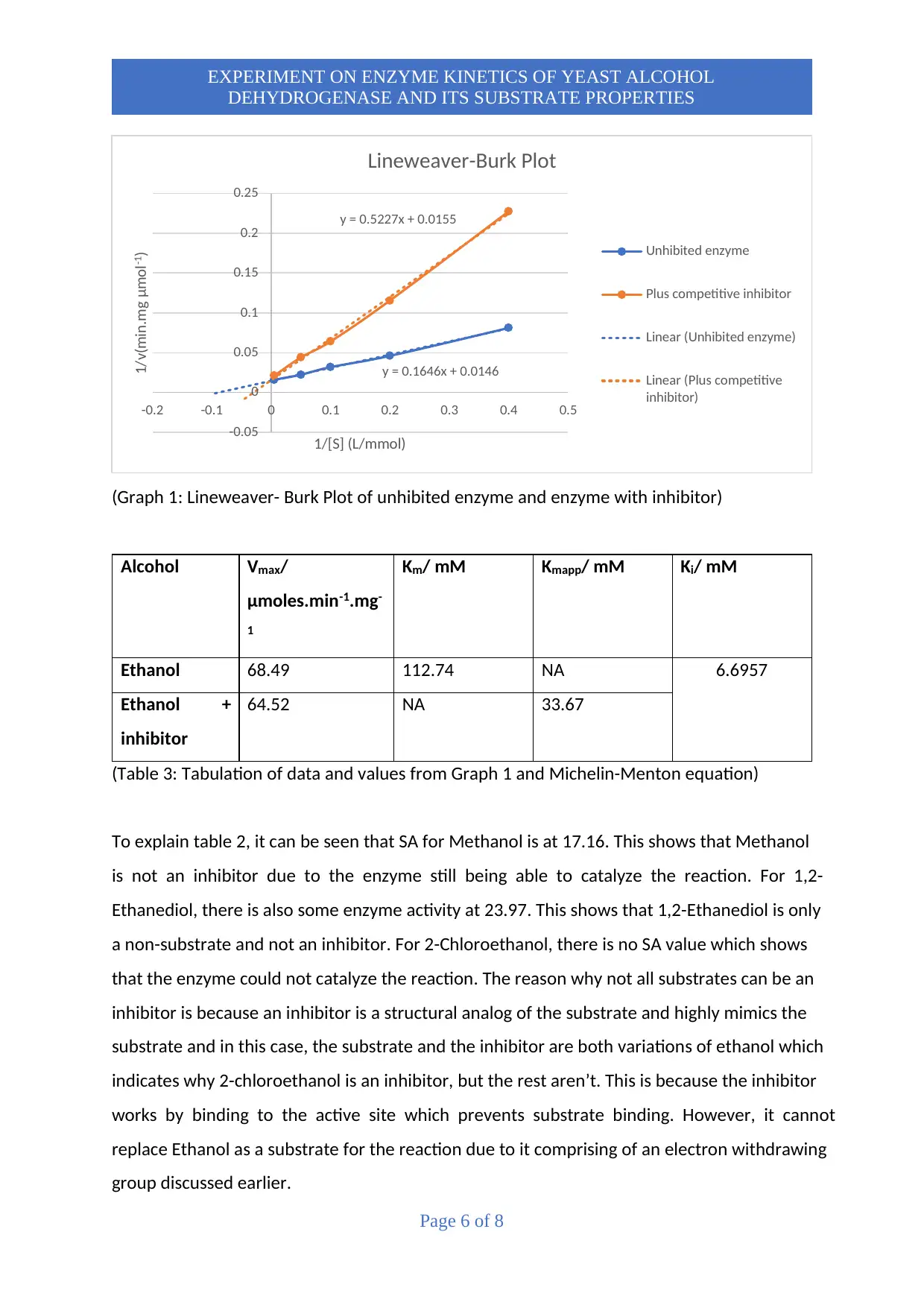
(Graph 1: Lineweaver- Burk Plot of unhibited enzyme and enzyme with inhibitor)
Alcohol Vmax/
μmoles.min-1.mg-
1
Km/ mM Kmapp/ mM Ki/ mM
Ethanol 68.49 112.74 NA 6.6957
Ethanol +
inhibitor
64.52 NA 33.67
(Table 3: Tabulation of data and values from Graph 1 and Michelin-Menton equation)
To explain table 2, it can be seen that SA for Methanol is at 17.16. This shows that Methanol
is not an inhibitor due to the enzyme still being able to catalyze the reaction. For 1,2-
Ethanediol, there is also some enzyme activity at 23.97. This shows that 1,2-Ethanediol is only
a non-substrate and not an inhibitor. For 2-Chloroethanol, there is no SA value which shows
that the enzyme could not catalyze the reaction. The reason why not all substrates can be an
inhibitor is because an inhibitor is a structural analog of the substrate and highly mimics the
substrate and in this case, the substrate and the inhibitor are both variations of ethanol which
indicates why 2-chloroethanol is an inhibitor, but the rest aren’t. This is because the inhibitor
works by binding to the active site which prevents substrate binding. However, it cannot
replace Ethanol as a substrate for the reaction due to it comprising of an electron withdrawing
group discussed earlier.
y = 0.1646x + 0.0146
y = 0.5227x + 0.0155
-0.05
0
0.05
0.1
0.15
0.2
0.25
-0.2 -0.1 0 0.1 0.2 0.3 0.4 0.5
1/v(min.mg μmol-1)
1/[S] (L/mmol)
Lineweaver-Burk Plot
Unhibited enzyme
Plus competitive inhibitor
Linear (Unhibited enzyme)
Linear (Plus competitive
inhibitor)
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
(Graph 1: Lineweaver- Burk Plot of unhibited enzyme and enzyme with inhibitor)
Alcohol Vmax/
μmoles.min-1.mg-
1
Km/ mM Kmapp/ mM Ki/ mM
Ethanol 68.49 112.74 NA 6.6957
Ethanol +
inhibitor
64.52 NA 33.67
(Table 3: Tabulation of data and values from Graph 1 and Michelin-Menton equation)
To explain table 2, it can be seen that SA for Methanol is at 17.16. This shows that Methanol
is not an inhibitor due to the enzyme still being able to catalyze the reaction. For 1,2-
Ethanediol, there is also some enzyme activity at 23.97. This shows that 1,2-Ethanediol is only
a non-substrate and not an inhibitor. For 2-Chloroethanol, there is no SA value which shows
that the enzyme could not catalyze the reaction. The reason why not all substrates can be an
inhibitor is because an inhibitor is a structural analog of the substrate and highly mimics the
substrate and in this case, the substrate and the inhibitor are both variations of ethanol which
indicates why 2-chloroethanol is an inhibitor, but the rest aren’t. This is because the inhibitor
works by binding to the active site which prevents substrate binding. However, it cannot
replace Ethanol as a substrate for the reaction due to it comprising of an electron withdrawing
group discussed earlier.
y = 0.1646x + 0.0146
y = 0.5227x + 0.0155
-0.05
0
0.05
0.1
0.15
0.2
0.25
-0.2 -0.1 0 0.1 0.2 0.3 0.4 0.5
1/v(min.mg μmol-1)
1/[S] (L/mmol)
Lineweaver-Burk Plot
Unhibited enzyme
Plus competitive inhibitor
Linear (Unhibited enzyme)
Linear (Plus competitive
inhibitor)
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

Page 7 of 8
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
Based on graph 1, it can be seen whereby as the concentration of substrate increases,
the enzyme activity also increases. This is because YAD enzyme functions more effectively
with higher substrate concentration to produce the enzyme substrate complexes due to more
substrate molecules being able to bind at the active site.
From the graph, it can be seen that with the competitive inhibitor, the enzyme activity
is lower as opposed to without using an inhibitor. This is expected due to the inhibitor
occupying the active sites. Theoretically, with a competitive inhibitor, the enzyme should not
function when the substrate is of low concentrations. However, in this experiment, even at
low concentrations, there is some minor enzyme activity. This could be due to the
concentration of the inhibitor used being very low at only 1mmol/L.
To understand further on the inhibition types, we have to study the Lineweaver-Burk
plot. As observed in Graph 1, the results show intersecting lines at y-axis for unhibited enzyme
and enzyme with inhibitor. This shows that it is a competitive inhibitor because the slope and
the x-intercept is affected. If an uncompetitive inhibitor was used, the lines in the graph will
be parallel whereas if a non-competitive inhibitor is used, the lines will not intersect in the y-
axis.
Based on graph 1, it has been calculated that with an inhibitor, Km app value is 33.67
whereas without an inhibitor, Km value is at 112.74. The lower the Km value, the higher the
enzyme affinity. Thus, it can be said that with an inhibitor, the enzyme affinity towards
ethanol molecule is higher which indicates stronger substrate binding. The Vmax for ethanol
is 68.49and for ethanol+inhibitor is 64.52. This shows that there is no significant difference in
Vmax because for competitive inhibitor, the inhibition action gives no effect at high substrate
concentration. Ki value is the inhibitory constant value. The smaller the Ki value, the greater
the binding affinity of the inhibitor. Thus, at 6.6957mM, it can be assumed that the inhibitor
is generally not that strong in binding affinity.
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
Based on graph 1, it can be seen whereby as the concentration of substrate increases,
the enzyme activity also increases. This is because YAD enzyme functions more effectively
with higher substrate concentration to produce the enzyme substrate complexes due to more
substrate molecules being able to bind at the active site.
From the graph, it can be seen that with the competitive inhibitor, the enzyme activity
is lower as opposed to without using an inhibitor. This is expected due to the inhibitor
occupying the active sites. Theoretically, with a competitive inhibitor, the enzyme should not
function when the substrate is of low concentrations. However, in this experiment, even at
low concentrations, there is some minor enzyme activity. This could be due to the
concentration of the inhibitor used being very low at only 1mmol/L.
To understand further on the inhibition types, we have to study the Lineweaver-Burk
plot. As observed in Graph 1, the results show intersecting lines at y-axis for unhibited enzyme
and enzyme with inhibitor. This shows that it is a competitive inhibitor because the slope and
the x-intercept is affected. If an uncompetitive inhibitor was used, the lines in the graph will
be parallel whereas if a non-competitive inhibitor is used, the lines will not intersect in the y-
axis.
Based on graph 1, it has been calculated that with an inhibitor, Km app value is 33.67
whereas without an inhibitor, Km value is at 112.74. The lower the Km value, the higher the
enzyme affinity. Thus, it can be said that with an inhibitor, the enzyme affinity towards
ethanol molecule is higher which indicates stronger substrate binding. The Vmax for ethanol
is 68.49and for ethanol+inhibitor is 64.52. This shows that there is no significant difference in
Vmax because for competitive inhibitor, the inhibition action gives no effect at high substrate
concentration. Ki value is the inhibitory constant value. The smaller the Ki value, the greater
the binding affinity of the inhibitor. Thus, at 6.6957mM, it can be assumed that the inhibitor
is generally not that strong in binding affinity.
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

Page 8 of 8
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
REFERENCES
Bennetzen J.L & Hall B.D, The primary structure of the Saccharomyces cerevisiae gene for
alcohol dehydrogenase. I. [Yeast]. The Journal of biological chemistry, 257, pp.The Journal of
biological chemistry, Vol.257.
DICKINSON, F. and P. MONGER, G., 1973. [online] Ncbi.nlm.nih.gov. Available at:
<https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1177466/pdf/biochemj00614-0097.pdf>
[Accessed 11 April 2021].
Leskovac, Vladimir, Trivić, Svetlana & Peričin, Draginja, 2002. The three zinc-containing
alcohol dehydrogenases from baker's yeast, Saccharomyces cerevisiae. FEMS yeast research,
2(4), pp.481–494.
Plapp, Bryce V, Charlier, Henry A & Ramaswamy, S, 2016. Mechanistic implications from
structures of yeast alcohol dehydrogenase complexed with coenzyme and an alcohol.
Archives of biochemistry and biophysics, 591, pp.35–42.
Raj, Savarimuthu Baskar, Ramaswamy, S & Plapp, Bryce V, 2014. Yeast Alcohol
Dehydrogenase Structure and Catalysis. Biochemistry (Easton), 53(36), pp.5791–5803.
Weinhold, E.G. et al., 1991. Structural determinants of stereospecificity in yeast alcohol
dehydrogenase. Proceedings of the National Academy of Sciences - PNAS, 88(19), pp.8420–
8424.
Wilcox, A. E, LoConte, Micaela A & Slade, Kristin M, 2016. Effects of Macromolecular Crowding
on Alcohol Dehydrogenase Activity Are Substrate-Dependent. Biochemistry (Easton), 55(25),
pp.3550–3558.
EXPERIMENT ON ENZYME KINETICS OF YEAST ALCOHOL
DEHYDROGENASE AND ITS SUBSTRATE PROPERTIES
REFERENCES
Bennetzen J.L & Hall B.D, The primary structure of the Saccharomyces cerevisiae gene for
alcohol dehydrogenase. I. [Yeast]. The Journal of biological chemistry, 257, pp.The Journal of
biological chemistry, Vol.257.
DICKINSON, F. and P. MONGER, G., 1973. [online] Ncbi.nlm.nih.gov. Available at:
<https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1177466/pdf/biochemj00614-0097.pdf>
[Accessed 11 April 2021].
Leskovac, Vladimir, Trivić, Svetlana & Peričin, Draginja, 2002. The three zinc-containing
alcohol dehydrogenases from baker's yeast, Saccharomyces cerevisiae. FEMS yeast research,
2(4), pp.481–494.
Plapp, Bryce V, Charlier, Henry A & Ramaswamy, S, 2016. Mechanistic implications from
structures of yeast alcohol dehydrogenase complexed with coenzyme and an alcohol.
Archives of biochemistry and biophysics, 591, pp.35–42.
Raj, Savarimuthu Baskar, Ramaswamy, S & Plapp, Bryce V, 2014. Yeast Alcohol
Dehydrogenase Structure and Catalysis. Biochemistry (Easton), 53(36), pp.5791–5803.
Weinhold, E.G. et al., 1991. Structural determinants of stereospecificity in yeast alcohol
dehydrogenase. Proceedings of the National Academy of Sciences - PNAS, 88(19), pp.8420–
8424.
Wilcox, A. E, LoConte, Micaela A & Slade, Kristin M, 2016. Effects of Macromolecular Crowding
on Alcohol Dehydrogenase Activity Are Substrate-Dependent. Biochemistry (Easton), 55(25),
pp.3550–3558.
1 out of 8
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2025 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.

