Analyzing Natural Selection's Role in Pathogen Genome Evolution
VerifiedAdded on 2023/03/17
|10
|2366
|92
Report
AI Summary
This report provides a comprehensive overview of the role of natural selection in the evolution of pathogen genomes. It begins by introducing natural selection models, including directional, balancing, and diversifying selection, and then delves into the tests used to identify natural selection, such as substitution rate patterns, dn/ds estimation, and the MacDonald Krietman Test. The report then examines the evolution of human pathogenic genomes, highlighting the increase in pathogens over time and the impact of diseases like HIV and HCV. Finally, it discusses the measures and initiatives taken by the UK in pathogen genomics, including the use of Whole Genome Sequencing techniques, the establishment of the National Infection Service, and the work done by the Public Health England and various research units. The report concludes by emphasizing the significance of natural selection in understanding and managing infectious diseases, and the importance of global awareness and initiatives.

1
ROLE OF NATURAL SELECTION IN THE EVOLUTION OF PATHOGEN
GENOMES
ROLE OF NATURAL SELECTION IN THE EVOLUTION OF PATHOGEN
GENOMES
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

2
Table of Content
Introduction................................................................................................................................3
Natural Selection Models...........................................................................................................3
Tests to identify Natural Selection.............................................................................................3
Pathogen genomics: measures and initiatives taken by UK.......................................................6
Conclusion..................................................................................................................................8
Reference List............................................................................................................................9
Table of Content
Introduction................................................................................................................................3
Natural Selection Models...........................................................................................................3
Tests to identify Natural Selection.............................................................................................3
Pathogen genomics: measures and initiatives taken by UK.......................................................6
Conclusion..................................................................................................................................8
Reference List............................................................................................................................9

3
Introduction
Human adaptation and other manifestations of Natural selection have helped researchers to
unravel various biological mechanisms of defiance to infectious diseases. Pathogens are
infections or infectious agents that cause fallibility and mortality among mankind and other
animals globally. While migrating all over the world, the human population have come across
some definite pathogens and natural selection has increased the pervasiveness of the
resistance alleles to evolve and spread over time. With the help of modern edge analytical
methods, advanced technology and extensive resources of public data, the natural selection is
unleashing some unique intuition regarding the functioning mechanism of human biology. In
this present study we will have an overall idea about how natural selection has affected the
evolution of Pathogen Genomes and what measures have been taken to resolve the infectious
diseases in UK through years.
Natural Selection Models
Natural selection is the process by which propitious genetic traits increase in abundance over
time, whereas the hostile genetic traits lose their abundance (Bentley & Parkhill, 2015). To
address this process several different models have been proposed for different values of
parameters concerned, like- mode of inheritance, tenacity of selection etc. (Biek et al., 2015)
All of the selection models possess two loci having two alleles and the selection takes place
on the basis of fitness among the 3 genotypes (one heterozygote and two homozygotes).
Directional Selection Model- until getting stability one of the two alleles is preferred
over the other and the abundance is also increased through time (Croll et al., 2015).
Balancing Selection Model- in this method a new allele has Quick and Stable
equilibrium frequency. Compared to both the homozygotes, the heterozygote has a
greater fitness (Delong et al., 2016).
Diversifying Selection Model- in this classical model, two or more phenotypes are
preferred simultaneously giving rise to an increased pattern in genetic variation (Croll
et al., 2015).
Tests to identify Natural Selection
Selection has different impacts on genetic variations. In a simple Directional Selection model,
novel mutation will be preferred if it is concerned about some selective advantages of the
pathogen genome (Dong, Raffaele & Kamoun, 2015). This phenomenon is termed as Positive
Introduction
Human adaptation and other manifestations of Natural selection have helped researchers to
unravel various biological mechanisms of defiance to infectious diseases. Pathogens are
infections or infectious agents that cause fallibility and mortality among mankind and other
animals globally. While migrating all over the world, the human population have come across
some definite pathogens and natural selection has increased the pervasiveness of the
resistance alleles to evolve and spread over time. With the help of modern edge analytical
methods, advanced technology and extensive resources of public data, the natural selection is
unleashing some unique intuition regarding the functioning mechanism of human biology. In
this present study we will have an overall idea about how natural selection has affected the
evolution of Pathogen Genomes and what measures have been taken to resolve the infectious
diseases in UK through years.
Natural Selection Models
Natural selection is the process by which propitious genetic traits increase in abundance over
time, whereas the hostile genetic traits lose their abundance (Bentley & Parkhill, 2015). To
address this process several different models have been proposed for different values of
parameters concerned, like- mode of inheritance, tenacity of selection etc. (Biek et al., 2015)
All of the selection models possess two loci having two alleles and the selection takes place
on the basis of fitness among the 3 genotypes (one heterozygote and two homozygotes).
Directional Selection Model- until getting stability one of the two alleles is preferred
over the other and the abundance is also increased through time (Croll et al., 2015).
Balancing Selection Model- in this method a new allele has Quick and Stable
equilibrium frequency. Compared to both the homozygotes, the heterozygote has a
greater fitness (Delong et al., 2016).
Diversifying Selection Model- in this classical model, two or more phenotypes are
preferred simultaneously giving rise to an increased pattern in genetic variation (Croll
et al., 2015).
Tests to identify Natural Selection
Selection has different impacts on genetic variations. In a simple Directional Selection model,
novel mutation will be preferred if it is concerned about some selective advantages of the
pathogen genome (Dong, Raffaele & Kamoun, 2015). This phenomenon is termed as Positive
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

4
Selection. Negative selection occurs if the mutation seems dangerous to the genome. Both the
selection processes can be measured on a single amino acid of the gene or across the whole
genome. 3 major procedures will be discussed further to infer selection methods in pathogen
genomes (Myers & Cory, 2016).
1. Substitution rate patterns- genes in microbial genomes generally mutate
independently with different populations. In response to common Selection Pressure,
a convergent or parallel evolution is identified by intermittent emergence of same
substitution within different individuals (Porse et al., 2016). During Tuberculosis
treatment, by antimicrobial drugs selective pressure is applied on the pathogen
Mycobacterium tuberculosis which is evident from the fact that the same drug
resistance point mutation within different bodies emerges frequently. The number of
substitution per gene can be easily calculated by multiplying the gene’s length and
mutation per site (Pugliese et al., 2016). Thus, any significant rise in the substitution
rate than from the expected value can be used as a clear indication of positive
selection.
2. dn/ds estimation- another popular method for the detection of selection between
divergent individuals is dn/ds estimation. dn stands for the rate of non-synonymous
substitution per non-synonymous site, where as ds is the counterpart for the
synonymous ones (Quainoo et al., 2017). In the absence of selection dn/ds ratio
remains zero, i.e, it signifies a neutral condition. If the value exceeds 1, it indicates a
positive selection and when the value is less than 1, it suggests a negative selection
(Sánchez et al., 2018). However, there are some restrictions while applying this model
for microbial populations (Croll et al., 2015). Firstly, if the expected number of
substitution at any individual is very small, then it will be difficult calculate the non-
neutral dn/ds. Secondly, due to higher existence of negative selection sites, the true
value of dn/ds goes below the expected value which reduces the statistical power of
positive selection site detection (Stukenbrock, 2016).
3. MacDonald Krietman Test- the MK test is the assessment for the ratio of Pn/Ps and
Dn/Ds. Pn/Ps is the ratio of non-synonymous to synonymous polymorphisms. Dn/Ds is
the ratio of non-synonymous to synonymous fixed contrasts (Stukenbrock, 2016). As
per the comment of Quainoo et al., (2017), this particular test is widely used to test
within species selection type but it can also be used to decipher selection among the
sub-populations. Dn/Ds> Pn/Ps indicates that the non-synonymous changes are higher
Selection. Negative selection occurs if the mutation seems dangerous to the genome. Both the
selection processes can be measured on a single amino acid of the gene or across the whole
genome. 3 major procedures will be discussed further to infer selection methods in pathogen
genomes (Myers & Cory, 2016).
1. Substitution rate patterns- genes in microbial genomes generally mutate
independently with different populations. In response to common Selection Pressure,
a convergent or parallel evolution is identified by intermittent emergence of same
substitution within different individuals (Porse et al., 2016). During Tuberculosis
treatment, by antimicrobial drugs selective pressure is applied on the pathogen
Mycobacterium tuberculosis which is evident from the fact that the same drug
resistance point mutation within different bodies emerges frequently. The number of
substitution per gene can be easily calculated by multiplying the gene’s length and
mutation per site (Pugliese et al., 2016). Thus, any significant rise in the substitution
rate than from the expected value can be used as a clear indication of positive
selection.
2. dn/ds estimation- another popular method for the detection of selection between
divergent individuals is dn/ds estimation. dn stands for the rate of non-synonymous
substitution per non-synonymous site, where as ds is the counterpart for the
synonymous ones (Quainoo et al., 2017). In the absence of selection dn/ds ratio
remains zero, i.e, it signifies a neutral condition. If the value exceeds 1, it indicates a
positive selection and when the value is less than 1, it suggests a negative selection
(Sánchez et al., 2018). However, there are some restrictions while applying this model
for microbial populations (Croll et al., 2015). Firstly, if the expected number of
substitution at any individual is very small, then it will be difficult calculate the non-
neutral dn/ds. Secondly, due to higher existence of negative selection sites, the true
value of dn/ds goes below the expected value which reduces the statistical power of
positive selection site detection (Stukenbrock, 2016).
3. MacDonald Krietman Test- the MK test is the assessment for the ratio of Pn/Ps and
Dn/Ds. Pn/Ps is the ratio of non-synonymous to synonymous polymorphisms. Dn/Ds is
the ratio of non-synonymous to synonymous fixed contrasts (Stukenbrock, 2016). As
per the comment of Quainoo et al., (2017), this particular test is widely used to test
within species selection type but it can also be used to decipher selection among the
sub-populations. Dn/Ds> Pn/Ps indicates that the non-synonymous changes are higher
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

5
than the fixed differences, suggesting a positive selective, while negative selection is
inferred when Dn/Ds< Pn/Ps (Croll et al., 2015).
Figure: Evolution of Human Pathogenic genomes
(Source: Quainoo et al., 2017)
Figure: origin of human pathogen
(Source: Pugliese et al., 2016)
From the age of modern humans, there has been a trend of increase in human pathogens as
well (Porse et al., 2016). Some pathogens are responsible for large human casualties, like- the
than the fixed differences, suggesting a positive selective, while negative selection is
inferred when Dn/Ds< Pn/Ps (Croll et al., 2015).
Figure: Evolution of Human Pathogenic genomes
(Source: Quainoo et al., 2017)
Figure: origin of human pathogen
(Source: Pugliese et al., 2016)
From the age of modern humans, there has been a trend of increase in human pathogens as
well (Porse et al., 2016). Some pathogens are responsible for large human casualties, like- the
6
bacteria Yersinia pestis causing Black Plague, the virus Variola responsible for Smallpox, the
Malaria protozoa Plasmodium etc. Just after the migration from Africa human lineage broke
into fragments both geographically as well as genetically (Dong, Raffaele & Kamoun, 2015).
Gradually, theses population started to mix through colonization and the population
increased. On closer contact with animals and people all over the world, infectious pathogens
became widespread. As a consequence, fatal diseases like, Malaria, Cholera, Smallpox,
Tuberculosis, Leprosy became more common. As the civilization is developing day by day,
mankind is facing more severe threats like HIV, AIDS, SARS (Myers & Cory, 2016).
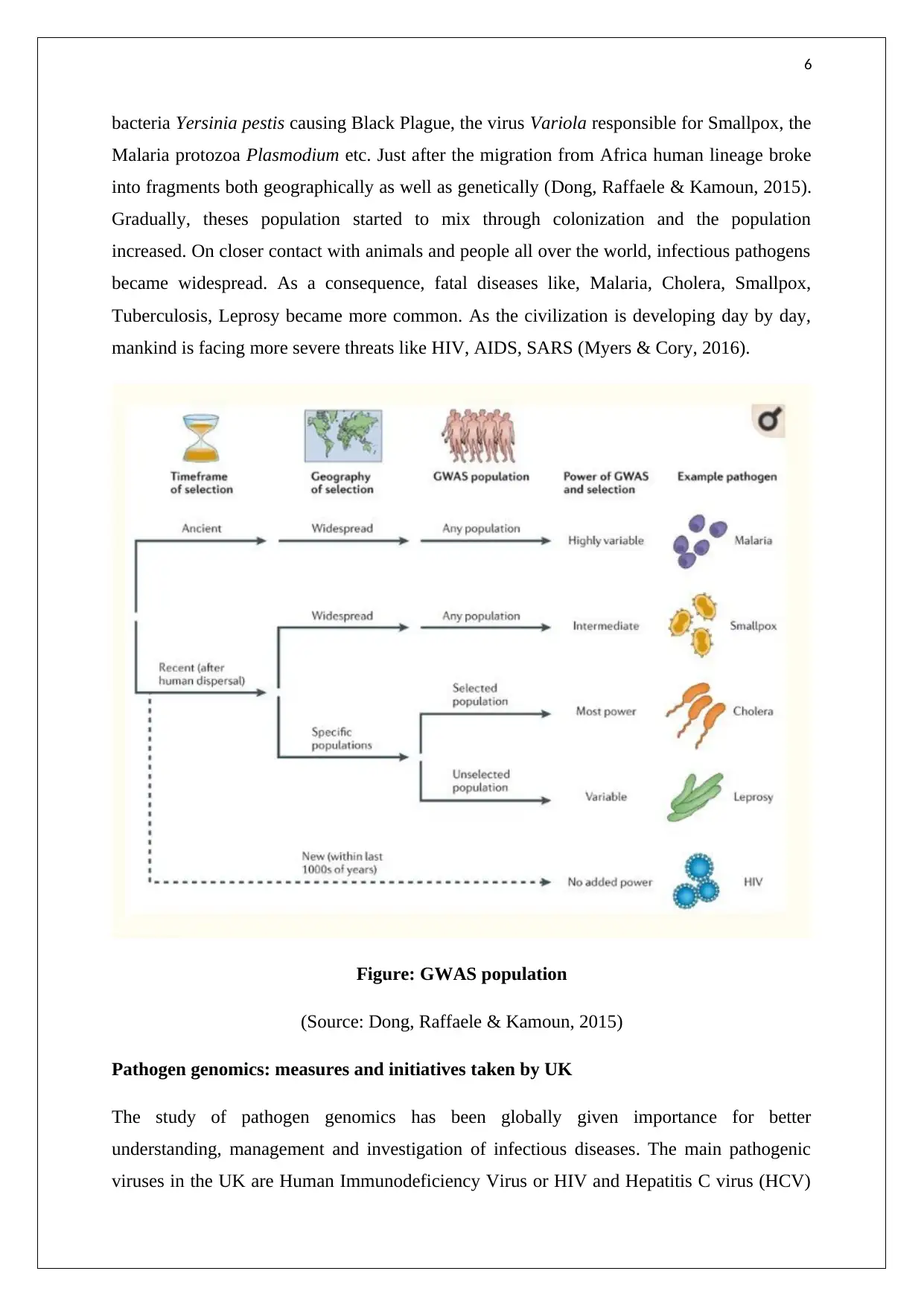
Figure: GWAS population
(Source: Dong, Raffaele & Kamoun, 2015)
Pathogen genomics: measures and initiatives taken by UK
The study of pathogen genomics has been globally given importance for better
understanding, management and investigation of infectious diseases. The main pathogenic
viruses in the UK are Human Immunodeficiency Virus or HIV and Hepatitis C virus (HCV)
bacteria Yersinia pestis causing Black Plague, the virus Variola responsible for Smallpox, the
Malaria protozoa Plasmodium etc. Just after the migration from Africa human lineage broke
into fragments both geographically as well as genetically (Dong, Raffaele & Kamoun, 2015).
Gradually, theses population started to mix through colonization and the population
increased. On closer contact with animals and people all over the world, infectious pathogens
became widespread. As a consequence, fatal diseases like, Malaria, Cholera, Smallpox,
Tuberculosis, Leprosy became more common. As the civilization is developing day by day,
mankind is facing more severe threats like HIV, AIDS, SARS (Myers & Cory, 2016).
Figure: GWAS population
(Source: Dong, Raffaele & Kamoun, 2015)
Pathogen genomics: measures and initiatives taken by UK
The study of pathogen genomics has been globally given importance for better
understanding, management and investigation of infectious diseases. The main pathogenic
viruses in the UK are Human Immunodeficiency Virus or HIV and Hepatitis C virus (HCV)
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

7
(Dong, Raffaele & Kamoun, 2015). HIV is responsible for the slow, progressive and most
fatal disease that is Acquired Immunodeficiency Syndrome or AIDS. Not only in UK, but
AIDS has become the most alarming global health burden nowadays, resulting almost a
million deaths worldwide within a year. Statistics reveal that annually around 500 people die
due to AIDS in UK. Medical survey states that in 2012, almost a million people of UK were
infected with HIV and among them 20-22% were not even aware of that fact (Dong, Raffaele
& Kamoun, 2015).
Whole Genome Sequencing techniques have been discovered to resolve the pathogen related
diseases. For this purpose, several initiatives have been taken and large number of
publications has been published. Chief Medical Officer’s Annual Report, 2016 is one of them
(Dong, Raffaele & Kamoun, 2015). European countries have taken some serious measures on
this particular issue. The Public Health England (PHE) has been trying to implement modern
WGS techniques in order to secure a healthy future. The National Infection Service (NIS) of
UK has a national strategy of WGS to intensify the control of pathogenic diseases and to help
PHE reach its goals in infectious disease management (Dong, Raffaele & Kamoun, 2015). At
PHE, “Colindale” a centralised WGS service has been established to provide services for a
range of nationally important pathogens including E. coli, Shigella, Listeria, Campylobacter,
S. aureus, Salmonella and Mycobacteria. The Colindale WGS service of UK was one of the
prime UK laboratories to get appraisal for its Whole Genome Sequencing services of virus
and bacteria. Among the initial adopters of the Whole Genome Sequencing approach for
conventional services delivery at PHE was the GBRU- Gastrointestinal Bacteria Reference
Unit. It was the National Reference laboratory for Gastrointestinal bacterial pathogens.
Gastrointestinal (GI) disease is the illnesses mainly caused by foodborne transmission of
bacteria or via being in direct touch with animals and their environments (Delong et al.,
2016). UCL has another pathogenic unit established by the grants from UK’s Medical
Research Council (MRC). The unit carries out the following:
i) Library preparation for whole genome sequencing (Delong et al., 2016)
ii) Library preparation for pathogen genome enrichment including clinical samples
iii) RNA sequencing for pathogen discovery
iv) Amplicon sequencing (Delong et al., 2016)
v) Viral resistance gene amplification
vi) Bioinformatics
vii) Sequence analysis (Delong et al., 2016)
(Dong, Raffaele & Kamoun, 2015). HIV is responsible for the slow, progressive and most
fatal disease that is Acquired Immunodeficiency Syndrome or AIDS. Not only in UK, but
AIDS has become the most alarming global health burden nowadays, resulting almost a
million deaths worldwide within a year. Statistics reveal that annually around 500 people die
due to AIDS in UK. Medical survey states that in 2012, almost a million people of UK were
infected with HIV and among them 20-22% were not even aware of that fact (Dong, Raffaele
& Kamoun, 2015).
Whole Genome Sequencing techniques have been discovered to resolve the pathogen related
diseases. For this purpose, several initiatives have been taken and large number of
publications has been published. Chief Medical Officer’s Annual Report, 2016 is one of them
(Dong, Raffaele & Kamoun, 2015). European countries have taken some serious measures on
this particular issue. The Public Health England (PHE) has been trying to implement modern
WGS techniques in order to secure a healthy future. The National Infection Service (NIS) of
UK has a national strategy of WGS to intensify the control of pathogenic diseases and to help
PHE reach its goals in infectious disease management (Dong, Raffaele & Kamoun, 2015). At
PHE, “Colindale” a centralised WGS service has been established to provide services for a
range of nationally important pathogens including E. coli, Shigella, Listeria, Campylobacter,
S. aureus, Salmonella and Mycobacteria. The Colindale WGS service of UK was one of the
prime UK laboratories to get appraisal for its Whole Genome Sequencing services of virus
and bacteria. Among the initial adopters of the Whole Genome Sequencing approach for
conventional services delivery at PHE was the GBRU- Gastrointestinal Bacteria Reference
Unit. It was the National Reference laboratory for Gastrointestinal bacterial pathogens.
Gastrointestinal (GI) disease is the illnesses mainly caused by foodborne transmission of
bacteria or via being in direct touch with animals and their environments (Delong et al.,
2016). UCL has another pathogenic unit established by the grants from UK’s Medical
Research Council (MRC). The unit carries out the following:
i) Library preparation for whole genome sequencing (Delong et al., 2016)
ii) Library preparation for pathogen genome enrichment including clinical samples
iii) RNA sequencing for pathogen discovery
iv) Amplicon sequencing (Delong et al., 2016)
v) Viral resistance gene amplification
vi) Bioinformatics
vii) Sequence analysis (Delong et al., 2016)
Paraphrase This Document
Need a fresh take? Get an instant paraphrase of this document with our AI Paraphraser

8
Conclusion
As pathogens’ genetic sequence changes over time, they are generally small and have a rapid
evolving rate; this is the only reason that they are effective and persistent agents of causing
infectious diseases. Natural selection plays a major role in investigating the evolution of such
pathogenic genomes. Scientists have discovered that ancient characteristics of balancing
selection along with the most recent characteristics of positive selection equally incriminate
the pathogens as one of the strongest selective causes of human evolution. Both the birth and
death rates are thus dependent on the evolution pathogenic genomes. UK has initiated some
good measures to deal with such infectious diseases carried by pathogenic genomes and has
also taken initiatives to spread awareness about the issue globally.
Conclusion
As pathogens’ genetic sequence changes over time, they are generally small and have a rapid
evolving rate; this is the only reason that they are effective and persistent agents of causing
infectious diseases. Natural selection plays a major role in investigating the evolution of such
pathogenic genomes. Scientists have discovered that ancient characteristics of balancing
selection along with the most recent characteristics of positive selection equally incriminate
the pathogens as one of the strongest selective causes of human evolution. Both the birth and
death rates are thus dependent on the evolution pathogenic genomes. UK has initiated some
good measures to deal with such infectious diseases carried by pathogenic genomes and has
also taken initiatives to spread awareness about the issue globally.

9
Reference List
Bentley, S. D., & Parkhill, J. (2015). Genomic perspectives on the evolution and spread of
bacterial pathogens. Proceedings of the Royal Society B: Biological Sciences, 282(1821),
20150488.
Biek, R., Pybus, O. G., Lloyd-Smith, J. O., & Didelot, X. (2015). Measurably evolving
pathogens in the genomic era. Trends in ecology & evolution, 30(6), 306-313.
Croll, D., Lendenmann, M. H., Stewart, E., & McDonald, B. A. (2015). The impact of
recombination hotspots on genome evolution of a fungal plant pathogen. Genetics, 201(3),
1213-1228.
Delong, T., Wiles, T. A., Baker, R. L., Bradley, B., Barbour, G., Reisdorph, R., ... & Elso, C.
M. (2016). Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide
fusion. Science, 351(6274), 711-714.
Dong, S., Raffaele, S., & Kamoun, S. (2015). The two-speed genomes of filamentous
pathogens: waltz with plants. Current opinion in genetics & development, 35, 57-65.
Myers, J. H., & Cory, J. S. (2016). Ecology and evolution of pathogens in natural populations
of Lepidoptera. Evolutionary applications, 9(1), 231-247.
Porse, A., Schønning, K., Munck, C., & Sommer, M. O. (2016). Survival and evolution of a
large multidrug resistance plasmid in new clinical bacterial hosts. Molecular biology and
evolution, 33(11), 2860-2873.
Pugliese, M., Loreto, V., Pompei, S., & Tria, F. (2016). Exploring the evolution of pathogens
organised in discrete antigenic clusters. Journal of Statistical Mechanics: Theory and
Experiment, 2016(9), 093306.
Quainoo, S., Coolen, J. P., van Hijum, S. A., Huynen, M. A., Melchers, W. J., van Schaik,
W., & Wertheim, H. F. (2017). Whole-genome sequencing of bacterial pathogens: the future
of nosocomial outbreak analysis. Clinical microbiology reviews, 30(4), 1015-1063.
Sánchez-Vallet, A., Fouché, S., Fudal, I., Hartmann, F. E., Soyer, J. L., Tellier, A., & Croll,
D. (2018). The genome biology of effector gene evolution in filamentous plant
pathogens. Annual review of phytopathology, 56, 21-40.
Reference List
Bentley, S. D., & Parkhill, J. (2015). Genomic perspectives on the evolution and spread of
bacterial pathogens. Proceedings of the Royal Society B: Biological Sciences, 282(1821),
20150488.
Biek, R., Pybus, O. G., Lloyd-Smith, J. O., & Didelot, X. (2015). Measurably evolving
pathogens in the genomic era. Trends in ecology & evolution, 30(6), 306-313.
Croll, D., Lendenmann, M. H., Stewart, E., & McDonald, B. A. (2015). The impact of
recombination hotspots on genome evolution of a fungal plant pathogen. Genetics, 201(3),
1213-1228.
Delong, T., Wiles, T. A., Baker, R. L., Bradley, B., Barbour, G., Reisdorph, R., ... & Elso, C.
M. (2016). Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide
fusion. Science, 351(6274), 711-714.
Dong, S., Raffaele, S., & Kamoun, S. (2015). The two-speed genomes of filamentous
pathogens: waltz with plants. Current opinion in genetics & development, 35, 57-65.
Myers, J. H., & Cory, J. S. (2016). Ecology and evolution of pathogens in natural populations
of Lepidoptera. Evolutionary applications, 9(1), 231-247.
Porse, A., Schønning, K., Munck, C., & Sommer, M. O. (2016). Survival and evolution of a
large multidrug resistance plasmid in new clinical bacterial hosts. Molecular biology and
evolution, 33(11), 2860-2873.
Pugliese, M., Loreto, V., Pompei, S., & Tria, F. (2016). Exploring the evolution of pathogens
organised in discrete antigenic clusters. Journal of Statistical Mechanics: Theory and
Experiment, 2016(9), 093306.
Quainoo, S., Coolen, J. P., van Hijum, S. A., Huynen, M. A., Melchers, W. J., van Schaik,
W., & Wertheim, H. F. (2017). Whole-genome sequencing of bacterial pathogens: the future
of nosocomial outbreak analysis. Clinical microbiology reviews, 30(4), 1015-1063.
Sánchez-Vallet, A., Fouché, S., Fudal, I., Hartmann, F. E., Soyer, J. L., Tellier, A., & Croll,
D. (2018). The genome biology of effector gene evolution in filamentous plant
pathogens. Annual review of phytopathology, 56, 21-40.
⊘ This is a preview!⊘
Do you want full access?
Subscribe today to unlock all pages.

Trusted by 1+ million students worldwide

10
Stukenbrock, E. H. (2016). The role of hybridization in the evolution and emergence of new
fungal plant pathogens. Phytopathology, 106(2), 104-112.
Stukenbrock, E. H. (2016). The role of hybridization in the evolution and emergence of new
fungal plant pathogens. Phytopathology, 106(2), 104-112.
Stukenbrock, E. H. (2016). The role of hybridization in the evolution and emergence of new
fungal plant pathogens. Phytopathology, 106(2), 104-112.
Stukenbrock, E. H. (2016). The role of hybridization in the evolution and emergence of new
fungal plant pathogens. Phytopathology, 106(2), 104-112.
1 out of 10
Related Documents
Your All-in-One AI-Powered Toolkit for Academic Success.
+13062052269
info@desklib.com
Available 24*7 on WhatsApp / Email
![[object Object]](/_next/static/media/star-bottom.7253800d.svg)
Unlock your academic potential
Copyright © 2020–2026 A2Z Services. All Rights Reserved. Developed and managed by ZUCOL.



